Introduction
The number of demands for fishery products has increased, as evidenced by the national fish consumption in 2019, which had an average of 54.50%. Fish commodities with high commercial value are: tuna, eastern little tuna, and skipjack tuna. Eastern little tuna has a complete nutritional content and has economic value. The production volume of the Scombridae group in 2021 reached 174.7 thousand tons and is distributed globally and domestically in the local market (BPS, 2021). The distribution process of fish must be carried out properly because fish is a perishable food. Therefore, it is easy for pathogenic and non-pathogenic microbes to grow. The correct temperature is required during the distribution process to avoid quality deterioration (Zhuang et al., 2021). The risk of shelf life and quality of fish products were determined by time and temperature in the cold chain (Lorentzen et al., 2022). The cold chain is part of a product processing system that aims to maintain product temperature during handling so as not to degrade its quality.
The stages in the cold chain need to consider the fish quality maintained with initial handling, processing to land, transportation, and storage of products before they reach consumers. The principle of the cold chain is to control the temperature of the fish to remain low, close to 0°C to maintain the quality of the fish during the distribution process. The application of low temperatures can be done using ice media. Storage temperature affects the chemical properties of fish such as histamine levels (Nurilmala et al., 2019), organoleptic physical properties (Lacapa et al., 2021) and the growth of bacteria and fungi in fish (Zhuang et al., 2021). The quality of eastern little tuna remains good at low temperatures because the formation of histamine by bacteria by changing histidine can be minimized. Histamine compounds are the cause of poisoning cases after consuming the eastern little tuna. Bacteria that produce the histidine decarboxylase enzyme as histamine-forming are inactive during cold temperatures (Norita et al., 2019). Tuna fish has an unsaturated fat that is easily oxidized, degraded, decomposed, and denatured, reducing fish quality.
Fish quality is also influenced by environmental factors such as temperature and handling. Tuna fish sold and handled in traditional markets and fish in modern markets have different qualities. Fish from traditional markets have a higher relative abundance and diversity of bacteria than fish from modern markets and might be affected by the low hygiene conditions of fish in traditional markets. Fish in traditional markets are sold in the open air without adding ice, and direct contact between the fish and the banana leaf is used as packaging. Handling fish in traditional markets does not use gloves so it can be a source of contamination. Fish sold in modern markets is sheltered with ice in glass containers, not exposed to outside air, and handled with high hygiene. This difference in handling methods is the cause of the differences in the diversity and community of bacteria found in fish from the two places. Low-temperature storage is the best way to prevent bacterial growth. Tuna stored at high temperatures (30°C) increased histamine concentrations at toxic levels (Arnold et al., 1980). The difference in cold temperature on storage affects the bacterial species that grow.
Bacterial communities growing at different temperatures can be analyzed using DNA samples to indicate chemical structure. The next step is that the DNA extract from the extraction process is amplified independently using a primer pair designed for the target DNA fragment and then sequenced. Bacterial community diversity is estimated using bioinformatics analysis. The results of the analysis were then identified through DNA molecular sequencing. DNA analysis using 16S ribosomal RNA (rRNA) sequences has been commonly used because it has high accuracy with a base length of 1,500 bp (Jandhyala et al., 2015). The 16S rRNA sequence has nine fragments, and the fragments used in this study were V3-V4. This fragment can increase alpha diversity, such as the Shannon and Simpson index values. The value of these two indices is the main factor in measuring the diversity of bacterial species in the sample. The results of this analysis indicate the bacterial community present in the product. The type of bacteria found in fish will determine the quality of the fish. Therefore, a cold chain system is needed to determine the effect of temperature on the bacterial community and the quality of eastern little tuna obtained from traditional markets. This study aims to determine the quality of eastern little tuna (Euthynnus affinis) and identify bacterial communities that grow on the surface of eastern little tuna (E.affinis) from traditional markets by applying a cold chain system.
Materials and Methods
Fish samples from Warung Jambu Main Market (Bantarjati, Bogor, West Java) were packaged in plastic and transported in fresh condition using ice in a cool box. This research started by collecting samples of 12 fresh eastern little tuna (E.affinis). The freshness of the fish was in good organoleptic condition, such as bright eyes, flat eyeballs, and clear corneas. The condition of the red gills is less bright and without mucus; on the surface of the fish, the mucus is clear and transparent with no color change. The fish texture is compact, elastic when pressed with a finger, and species-specific odor. Fish are transported to the laboratory using a cold chain system. Fish were transported in a cool box filled with ice at 4°C. The first step after the samples arrived at the laboratory was the initial pH measurement of all fish samples. Each eastern little tuna was put in a plastic bag—one fish as a sample without treatment (control). The plastic bags containing fish were put into three boxes labeled “contains ice” and coded A, B, and C. Each box was filled with three fish bags. Fish samples in boxes A, B, and C were stored for 24 hours in cold conditions at temperatures of 2°C, 4°C, and 10°C, respectively. Storage temperature is maintained by observing and checking every 2 hours gradually. Ice was added if the temperature increases from the specified temperature. The cold storage treatment diagram for fish samples can be seen in Fig. 1.
After the fish were treated, the surface of the eastern little tuna (approx. 390 mm × 100 mm) was swabbed using a sterile cotton swab (ONEMED, Krian, Indonesia) to obtain bacterial DNA. This process is carried out by gently rotating the head of a sterile cotton swab and rubbing the surface of the fish skin vertically, horizontally, and diagonally. The results of the swab on a sterile cotton swab were inserted into each culture glass tube which already contains 5 mL of 0.5% (w/v) nutrien broth (NB), by breaking the stem of the cotton swab sterile. The results of the swab in the solution were incubated.
The results of the eastern little tuna surface swab were then incubated in 0.5% (w/v) NB solution. The screw glass tube containing the swab resulted in 0.5% NB solution and was incubated using a water bath shaker for 24 hours at 31°C. Incubation at a temperature of 31°C adjusted to seawater temperature conditions. The incubation results were mixed using a vortex before being transferred into a 1.5 mL microtube and then centrifuged at a speed of 17,000×g for 10 minutes at a temperature of 10°C. The supernatant was discarded, and pellets were obtained as bacterial biomass, which was then used for DNA isolation.
Bacterial DNA isolation was performed using the commercial Qiagen DNeasy Blood & Tissue Kits (Qiagen, Germantown, MD, USA). The isolation technique uses bacterial biomass that has been obtained from the previous incubation process. DNA isolates were stored at –20°C before being used in the following analysis.
Next generation sequencing (NGS) technology was used for sequencing DNA isolates with good concentration and purity. DNA isolates with good concentration and purity had criteria such as 1.8–2.0 purity, no contamination or degradation, and a concentration of 20 ng/μL. DNA isolates that meet these criteria were sequenced with the Illumina NovaSeq 6000 (Illumina Inch, San Diego, CA, USA) platform. The specifications are read length with paired-end 250 bp sequencing depth up to 100,000 raw tags and sequence data quality 75% base with Q30 or higher. The target of the bacterial 16S rRNA gene was the primer used in the amplification process. The variable region used is V3-V4, with a forward primer 34F:5’CCTAYGGGRBGCASCAG and a reverse primer 860R:5’GGACTACNNGGGTATCTAAT.
The water activity (aw) measurement on the sample was carried out using an AQUALAB 4TE aw meter (METER Group Inch, Pullman, WA, USA). The sample was put into the measuring container provided in the tool. The eastern little tuna sample was inserted into the sample cup with no more than half the volume of the sample cup (7.5 mL). The sample was placed in a chamber, closed, and after 5 minutes of the reading process results appeared on the screen.
The value of drip loss was obtained from the shrinkage due to the discharge of fish flesh. Measurements were made by weighing the fish, plastic packaging, and the water in fish flesh that came out during treatment (w1). The weighing results were reduced by the results of weighing fish and packaging. The value obtained is divided by subtracting w1 by the weight of the fish packaging. The formula used was as follows.
W = drip loss value (%)
w1 = weight of packaging, water in fish flesh, and fish (gram)
w2 = weight of packaging and fish (gram)
w3 = weight of packaging (gram)
NGS data was analyzed quantitatively and translated into useful information using Bioinformatics software. Metagenomic analysis was used to determine the microbial community in the sample in the form of taxonomic classification. Taxonomic classification information is determined based on the 16S rRNA gene sequence. The sequence data obtained were then analyzed using Quantitative Insights into Microbial Ecology (QIIME) 2 software. QIIME 2 is a microbiome bioinformatics platform for analyzing raw DNA sequence data with decentralized provenance data and visualizing that provide new perspectives. The results obtained are in the form of figures and statistical results with publication-quality (Bolyen et al., 2019). The analysis used the following pipeline: the cutadapt plug-in was used to clean the NGS sequence data from the primer (Martin, 2011). This cleaning process is done with the demux-paired command, quality filtering, and denoising sequences using the DADA2 plug-in (Callahan et al., 2016). This step uses the denoised-paired command and the max ee 1.5 parameter minimum sequence above 200 bp; feature classifier (Bokulich et al., 2018). The commands used are classified sklearn and SILVA v138 database (Quast et al., 2013). The SILVA database provides datasets of aligned small and large subunit rRNA sequences for Bacteria, Archaea, and Eukarya. The datasets have updated, and quality checked regularly (Quast et al., 2013). The R program (R Core Team, 2013) was used for the analysis of alpha and beta diversity through Rstudio (R Studio team, 2020) and the Ampvis 2 package (Andersen et al., 2018).
The effect of cold chain system simulation on fish quality, including parameters of aw value, pH, and drip loss, was analyzed using the Statistical Package for the Sciences Social (SPSS) software program. The study was conducted in a completely randomized design (CRD) with three levels of storage temperature treatment, namely 2°C, 4°C, and 10°C, with three replications for each type of fish. Data analysis that has been distributed normally and homogeneously is analyzed by analysis of variance (ANOVA) or one-way ANOVA. The CRD test model is as follows.
Yij = score
µ = grand mean
αi = the effect of storage temperature treatment i, 2°C, 4°C, and 10°C
εij = the residual or random error
The hypothesis used is as follows:
H0: Different cold temperature treatments on fish storage did not affect the changes in the quality parameters of the fish produced.
H1: Different cold temperature treatments in fish storage affect changes in the quality parameters of the fish produced.
A significant effect was stated if the data analysis shows a 95% confidence interval (α = 0.05). If the results showed a significant difference (p < 0.05), further tests were carried out using Duncan’s. Duncan’s multiple range test was carried out with the following formula:
Rp = critical value for the treatment being compared
r = Duncan’s significant range value
α = significance level 5%
p = compared treatment or range value
dbs = degree of-freedom
KTS = middle square
Results
The data from the research that has been done shows a change in the average pH value of the fish that were not treated cold (control) and given cold treatment. Changes in the pH value of the sample can be seen in Table 1. The average pH value obtained in the control sample was 6.18. The average pH values obtained in samples A2, A4, and A10 were 5.93, 6.10, and 6.22, respectively (Table 1). The analysis of variance showed that cold storage was not significantly different from changes in pH value (p-value > 0.05).
| Storage treatment | pH value | Comparison data |
|---|---|---|
| Control | 6.18 ± 0.01a | 6.021) |
| A2 | 5.93 ± 0.14ab | 5.742) |
| A4 | 6.10 ± 0.10ab | 6.243) |
| A10 | 6.22 ± 0.11a | 6.254) |
The eastern little tuna sample A2, storage treatment at 2°C; A4, storage treatment at 4°C; A10, storage treatment at 10°C.
a,b The same superscript letter in the same column means that the analysis of variance test results are not significantly different (α = 0.05). n = repetition.
1) Data from Norita et al. (2019).
2) Data from Pianusa et al. (2015).
3) Data from Wiranata et al. (2017).
4) Data from Sari et al. (2018). n = 3.
The aw value obtained in Table 2 shows that the number of water microbes used in the sample is around 0.89 to 0.93 from the maximum value of aw is 1. The variance analysis results show that cold storage is not significantly different from changes in the aw value (p-value > 0.05).
| Storage treatment | Value of aw (n = 3) |
|---|---|
| Control | 0.907 ± 0.04a |
| A2 | 0.898 ± 0.03ab |
| A4 | 0.903 ± 0.01b |
| A10 | 0.932 ± 0.05ab |
The eastern little tuna sample A2, storage treatment at 2°C; A4, storage treatment at 4°C; A10, storage treatment at 10°C.
The value of the sample drip loss can be seen in Table 3. The results in Table 3 show that the lowest drip loss percentage is in the sample with a temperature treatment of 2°C, which is about 4.58%. Samples with 10°C treatment had the highest drip loss value, about 5.66%. The results of the analysis of variance showed that cold storage was not significantly different from changes in the value of drip loss.
| Storage treatment | Drip loss (%) |
|---|---|
| A2 | 4.58 ± 1.63a |
| A4 | 5.35 ± 0.74a |
| A10 | 5.66 ± 1.91ab |
The comparison of raw data sequences between A2, A10, and C1 research can be seen in Table 4. Table 4 shows that the raw read in sample A2 is 135,206 sequences, A10 is 179,839 sequences, and C1 is 187,101 sequences. The number of bacterial DNA sequences in sample C1 significantly differs from samples A2 and A10.
| Storage treatment | Input | Filtered | Denoised | Chimeric |
|---|---|---|---|---|
| A2 | 135,206 | 109,381 | 108,513 | 16.61 |
| A10 | 179,839 | 136,615 | 134,888 | 22.95 |
| C1 | 187,101 | 146,137 | 144,158 | 40.61 |
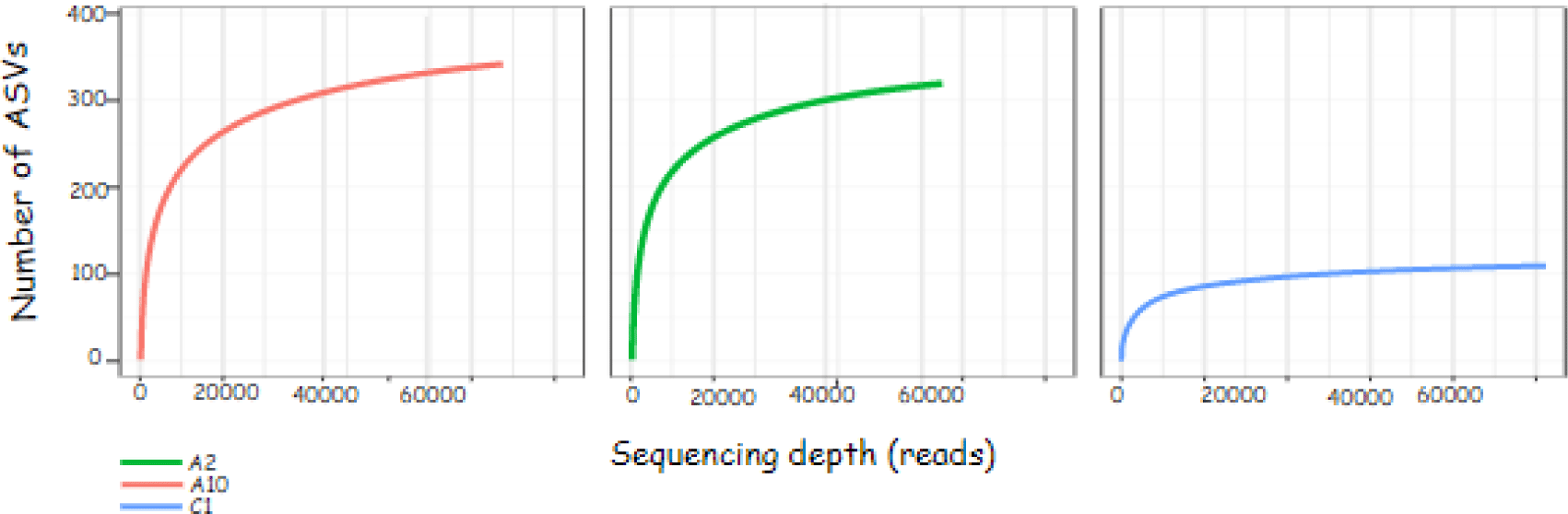
The sequence results obtained can be described through the rarefaction curve. The rarefaction curve in Fig. 2 shows the sequencing effort carried out on the sample is quite good. The flatter graph shows the sequencing depth is sufficient to identify all amplicon sequence variants (ASVs) in the sample. ASV values are the new name for operational taxonomic units.
This study used Alpha diversity to measure the diversity of the bacterial community in the sample based on 16S rRNA sequencing data. The results of the Alpha diversity index measurement can be seen in Table 5. Table 5 shows the observed ASVs in samples A2 and A10, respectively 325 and 319. The shannon index value in samples A2 and A10, respectively 2.35 and 3.08. The simpsons index value in samples A2 and A10, respectively 0.83 and 0.81.
| Storage treatment | Observed ASVs | Shannon index | Simpsons index |
|---|---|---|---|
| A2 | 325 | 2.35 | 0.83 |
| A10 | 319 | 3.08 | 0.81 |
| C1 | 104 | 3.18 | 0.16 |
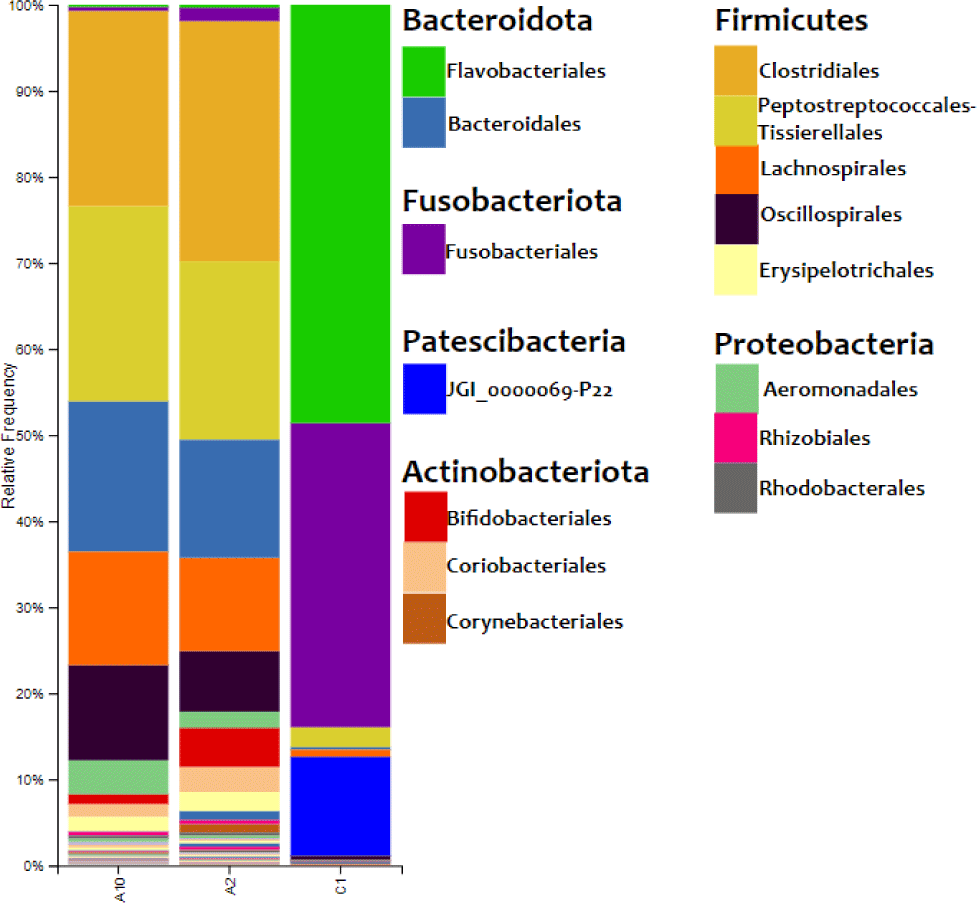
The results of the bacterial DNA sequencing process could describe the composition of bacteria on the surface of eastern little tuna. The relative abundance of bacterial communities in eastern little tuna samples A2, A10, and C1 at the order level can be seen in Fig. 3. The bar chart of bacterial communities in Fig. 3 shows that the phyla identified in samples A2 and A10 were 6 phyla, while C2 was 8 phyla. Firmicutes and Bacteridota are phyla with the highest percentage identified in samples A2 and A10. The diagram shows the order Clostridiales from the Firmicutes phylum dominates in samples A2 and A10 with percentages of 27.897% and 22.462%, respectively, while the dominant order in C1 is Flavobacteriales 48.603%. The orders of bacteria with a reasonably high abundance in sample A2 were Lachnospirales (10.817%), Oscillospirales (7.050%). As for sample A10, the same order was also found with the percentage of Lachnospirales (13.119%) and Oscillospirales (11.058%).
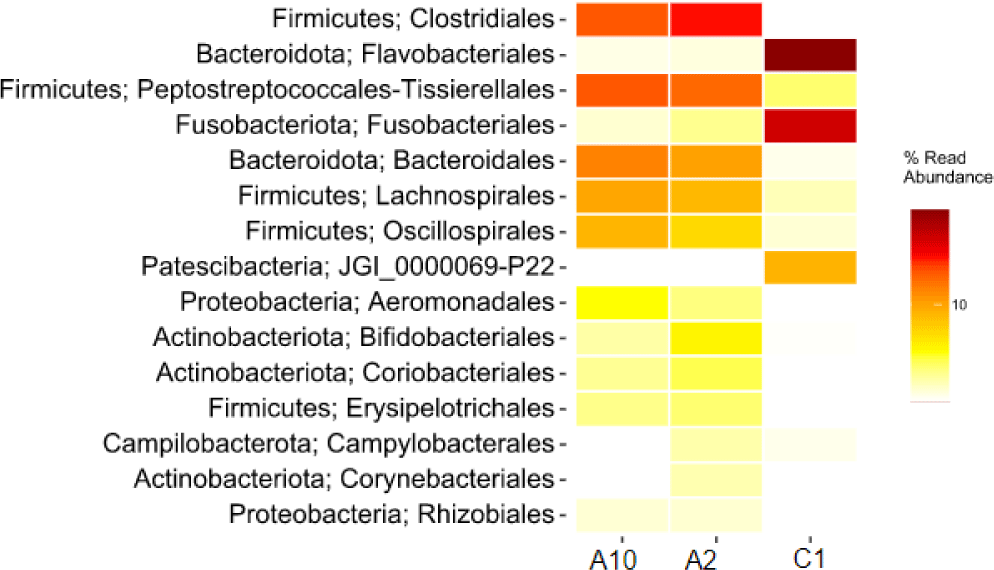
The heatmap of bacteria in order level in eastern little tuna samples A2, A10, and C1 at the order level can be seen in Fig. 4. The orders Clostridiales and Peptostreptococcales-Tissierellales had the highest abundance in samples A2 and A10. The order of bacteria with the highest abundance in sample C1 was Flavobacteriales; this can be seen from the color displayed on the order which has a high concentration on the heatmap visualization. The order Clostridiales in samples A2 and A10 had an abundance of 27.9% and 22.6%, respectively. The order Peptostreptococcales-Tissierellales in samples A2 and A10 had an abundance of 20.7% and 22.7%, respectively. Sample C1 with an abundance of the order Flavobacteriales 48.6%. Fig. 4 shows that the types and abundance of bacteria in samples A2 and A10 differ from C1. The dominant orders in samples A2 and A10 were Clostridiales and Peptostreptococcales-Tissierellales, only found with a relative abundance of 0%–2.3% in sample C1. The dominant order in sample C1 was Flavobacteriales, also identified in samples A2 and A10, with a relative abundance ranging from 0.3% to 0.4%.
The data obtained from the sequencing results show that several orders of bacteria found in samples A2 and A10 can potentially spoil fish and pathogens for humans who consume them. The order of spoilage bacteria causes physical damage. The types of bacteria identified as potential pathogens and spoilage in samples A2 and A10, as well as in sample C1, can be seen in Table 6. Table 6 shows that the pathogenic bacteria identified in the sample have a relatively low abundance in the 0.1%–0.7% range. In the samples, decomposing bacteria such as Lysinibacillus xylanilyticus, Dickeya phage, and Acinetobacter were in the range of 0.1%–1.4%.
Discussion
The level of freshness of fish can be observed from the degree of acidity or pH value. Fish with different storage treatments had different pH values due to the autolysis process and growing bacteria. Bacterial growth resulting from changes in pH is closely related to determining the quality of fish meat. Storage time and duration are factors that affect changes in pH during cooling. Research conducted by Nurilmala et al. (2019) showed that changes in pH during cold storage began to change on day 3. Storage carried out during this study was 24 hours, so this was the cause of insignificant changes in pH. Storage temperature is known to affect pH, as evidenced by storage at temperatures of 2°C and 4°C, which can reduce the pH value of fish compared to controls.
A comparison of the data obtained from this study with data by other researchers showed the same decreasing phenomenon at storage temperatures of 2°C and 4°C. Storage with a temperature range between 0°C–5°C reduces enzyme activity in fish meat, while high temperatures increase the pH value. Handling and transporting fish in summer has been shown to increase the acidity of fish. Low pH values reduce microbial activity due to the reduced activity of enzymes that play a role in the autolysis process (e.g., cathepsin) and spoilage in fish meat. The chilling temperature can inhibit the cathepsin enzyme’s activity, so the decay process can be controlled. Low pH values can also reduce bacteria’s ability to grow and prevent the breakdown of proteins into amino acids and volatile base compounds in fish meat by bacteria, thereby preventing alkaline conditions. An alkaline pH value greater than 7 indicates that the fish meat is not fresh.
aw in foodstuffs shows the free water content contained. aw indicates the amount of water that microbes can use to reproduce. Temperatures below the freezing point lower the vapor pressure of water and ice, lowering aw value. aw can be used as one of the parameters for determining food quality from the aspect of the types of microbes that grow and the chemical and biochemical reactions that occur. Each type of microbe requires a different value of water content to grow. Foodstuffs with an aw value greater than 0.98 are of particular concern because they provide suitable conditions for spoilage bacteria to grow. The aw value above 0.98 is a suitable habitat for the growth of Pseudomonas spp. (Harfan et al., 2019). These bacteria act as spoilage agents in foods with high protein content, such as fish stored at cold temperatures. Samples with an aw value of 0.89–0.97 became good conditions for the growth of gram-positive bacteria (especially the cocci group) and yeasts.
Changes in environmental temperature result in fluid release from the fish’s body, which is drip loss. The fluid that comes out contains several nutritional components that cannot be reabsorbed. The nutrients wasted in this fluid are included in the category of water-soluble sarcoplasmic proteins (Kaale et al., 2014). The reduced amount of protein is caused by the open structure of the meat so that it is unable to bind water and causes the value of drip loss to increase. The length of storage time is a factor that affects changes in drip loss during cooling. The difference in the value of drip loss obtained is influenced by the temperature used during storage. The temperature difference will affect the strength of the connective tissue and fat tissue to unite the fibers in fish muscle tissue and the branching between muscle fibers. Low temperatures prevent the interfibrillar space from stretching so that drip loss can be avoided (Jiang et al., 2020). Liquids containing nutrients such as protein cannot be reabsorbed by fish body tissues, thereby reducing the quality of the fish (Meiriza et al., 2016).
Analysis of sample sequence data can be performed using the QIIME 2 software (Bolyen et al., 2019). The primers on the NGS sequence data were cleaned, then the quality filtering and denoising processes were carried out using a plug-in in DADA 2 (Callahan et al., 2016). The raw read is carried out by a quality filtering and denoising process, namely, removing low-quality regions to improve the sequence quality (Min et al., 2021). Differences in treatment in handling fish samples affect the sequence results. Temperature significantly affects the number of bacterial DNA sequences, which indicates the abundance of bacteria in the sample. The number of bacterial DNA sequences in sample C1 significantly differs from samples A2 and A10. Differences in temperature, environmental hygiene conditions, and sampling time cause the high bacterial sequences found in sample C1. Sample C1 from the study of analyzed eastern little tuna at 30°C. Relative humidity from the market and fish directly exposed to air and vehicle fumes lead to the development of bacteria in fish. A temperature of 30°C is an excellent condition for bacterial growth. This is evidenced by research data from Siburian et al. (2012) who found a much greater number of bacteria in milkfish stored at 30°C than at 10°C. Milkfish stored at 10°C did not find bacterial growth. Storage of fish at low temperatures can inhibit bacterial growth thereby maintaining fish quality.
The rarefaction curve contains information on the sequencing depth (reads) on the x-axis and the number of observed ASVs on the y-axis. The number of ASV indicates the diversity of bacterial species present in the sample. The principle of bacterial community analysis using ASV values is based on the level of sequence difference, for example 93%, 95% and 97% (Schloss, 2021). The flat curve indicates that the ASV identification with the sequencing effort is sufficient. This is evidenced by the number of ASVs that do not increase with the addition of sequencing depth (reads).
Measurement of diversity using Alpha diversity describes the structure of the ecological community related to richness (number of taxonomic groups) and evenness (distribution of the abundance of taxonomic groups; Willis, 2019). The alpha diversity index measurement results were able to describe the abundance of a community in the sample, including information on wealth (richness) and evenness (evenness). Richness and evenness values indicate the number and distribution of species evenly in the sample (Robert, 2019). These two values are the main factors in measuring the diversity of the bacterial community in the sample. The higher the Shannon index value, the higher the diversity of the bacterial community. The interpretation of the Simpsons index value is slightly different from the Shannon value, and this value has a range of 0–1. The Simpsons value close to 0 indicates that the sample has a higher level of diversity in the bacterial community. The results of the diversity index measurement in the table above show that sample A10 has a higher level of diversity in the bacterial community than sample A2 due to the temperature treatment applied. The results also evidence this on sample C1 with a temperature treatment of 30°C. Sample C1 has a higher Shannon value and a lower Simpson value. Thus, concludes that sample C1 has a higher diversity of bacteria than samples A2 and A10. The diversity of the bacterial community increases if the species richness and evenness of the community increase in a sample (Kim et al., 2017). The Shannon index value describes the richness of the species, while Simpson’s index describes evenness.
The results of the relative abundance of bacteria in the eastern little tuna sample in Fig. 3 show Firmicutes and Bacteriodota are phyla with the highest percentage identified in samples A2 and A10. Research conducted by Carda-Diéguez et al. (2014) found the same data: the phylum Firmicutes, Fusobacteria, and Proteobacteria were the dominant bacteria found in Anguilla anguilla fish. These results were supported by data from research by Rimoldi et al. (2020), who found the dominant bacteria in European sea bass (Dicentrarchus labrax Linnaeus, 1758), one of which came from the phylum Firmicutes.
The order Clostridiales from phylum Firmiutes dominates in samples A2 and A10. The dominance of the Clostridiales order in this sample is supported by data from Rodríguez et al. (2021) proving that this type of bacteria is abundant in marine waters. This is the cause of the dominance of the order Clostrdiales bacteria in the sample. Apart from their habitat, the process of handling fish while on board and the ice used to freeze fish are also sources of bacterial contamination (Zhuang et al., 2021). Fish come into direct contact with ice cubes, the raw material for which comes from sea water, for a long time. Bacterial types of Clostridia sp., Micrococcus, Lactobacillus, Leuconostoc and Tetragenococcus are gram-positive bacteria that form histamine in tuna and skipjack. The presence of bacteria from the order Closriadiales can potentially cause high histamine levels in fish. Low temperature storage can inhibit the growth of histamine-forming bacteria so that it can keep histamine levels in fish below safe limits (Nurilmala et al., 2020).
Lachnospirales bacteria were found in both samples A2 and A10. Lachnospirales bacteria are gram-stain-positive, highly anaerobic, nonmotile, non-spore-forming, and spindle-shaped rods arranged in pairs and short chains. These bacteria were able to synthesize lactic, formic, acetic, and succinic acids as end products of fermentation from glucose. The quality of the eastern little tuna samples was influenced by other bacteria identified with low abundances, such as the orders Bacteroidota, Actinobacteriota, and Fusobacteria. The order Bacteroides is commonly found in marine waters, on the skin, and intestines of animals. These bacteria do not form spores, are rod-shaped, and include gram-negative bacteria. Research conducted by Zhang et al. (2021) also found bacteria of the order Fusobacteria in Zebrafish (Danio rerio Hamilton, 1822). This order of bacteria can help metabolize carbohydrates such as mucin into short-chain fat granules that can add energy to gastrointestinal cells and inhibit potential pathogens of freshwater fish (Rimoldi et al., 2018).
The bacterial community found in the eastern little tuna at the same sampling site may show different results. This study found the order of bacteria Flavobacteriales in the eastern little tuna without cold treatment obtained from traditional markets. The percentage of the relative abundance of Flavobacteriales in the sample at 30°C (C1) was 48.603% (Fig. 3).
Visualization of differences in the relative abundance of bacteria in the three samples can also be presented through a heatmap which is shown in the form of different color gradations. Visualizing the heatmap makes it easy to compare the relative abundance of bacteria in the three samples. The heatmap of the relative abundance of bacteria at the order level in Fig. 4 shows the difference in the abundance of bacteria of an order of bacteria in each different sample. This result can be influenced by sampling by researchers with different hygiene conditions. The handling process, equipment cleanliness of equipment, and surfaces that come into contact with the fish are factors in the results found. Flavobacteriales bacteria is one type of spoilage bacteria species in fish (Shehata et al., 2020).
These bacteria are commonly found in natural aquatic environments and aquariums. The disease caused by these bacteria is characterized by the appearance of an external infection on the surface of the fish’s body, gills, or fins. In fish that are still alive, the disease often ends in death, which causes huge economic losses in the aquaculture industry (Jumria et al., 2017). Cold storage temperature treatment of samples A2 and A10 was able to suppress the growth of spoilage bacteria, for example the order Flavobacteriales to maintain fresh fish morphology.
The bacterial genus that has potential as a group of spoilage and pathogenic bacteria was also identified in the sample. The genus that has the potential as pathogenic bacteria include Proteus and Photobacterium. Bacteria from the genus Proteus are gram-negative and are capable of causing injuries to the fins and infecting the muscle tissue of fish. The genus Proteus is found in both humans and animals (Drzewiecka, 2016). Proteus mirabilis and Proteus haussii bacteria have become a new threat to aquaculture and cause high mortality in several types of fish, for example, carp (Cyprinus carpio Linnaeus, 1758) and grouper (Epinephelus akaara Temminck & Schlegel, 1842; Liu et al., 2016). The other pathogenic bacteria are Photobacterium: a gram-negative, oxidase-positive, and catalase-positive bacteria in the family Vibrionaceae. These bacteria can produce the enzyme histidine decarboxylase to produce histamine. Histamine production by bacteria is affected by temperature, at 4°C and 30°C, respectively; histamine produced in tuna samples was 296 mg/kg and more than 2,000 mg/kg (Kanki et al., 2007).
Genus of bacteria that have the potential as spoilage bacteria includes L.xylanilyticus, D.phage, and Acinetobacter. L. xylanilyticus is a gram-positive, aerobic, spore-forming bacteria that grows optimally at pH 5–9 and a temperature range of 20°C–45°C (mesophiles). These bacteria can degrade xylan, which is a polymer unit b-1,4-D-xylopyranosil. Research by Wang et al. (2010) found bacterial isolates from the same genus as this bacterium, namely Lysinibacillus fusiformis, in the liver of puffer fish (Fugu obscurus Abe, 1949). This bacterium is known to have the ability to produce tetrodotoxin. Tetrodotoxin (TTX; C11H17N3O8, molecular weight 319) is a potent neurotoxin that causes paralysis by attacking nerves and leading to death (Hinman & Du Bois, 2003).
The low relative abundance of pathogenic and spoilage bacteria groups indicates that the eastern little tuna samples from markets A2, A10, and C1 are safe for consumption. The traded tuna can maintain its quality and food safety by handling fish properly. One of the factors is the cold chain distribution applied to the sample. The transportation can be by car/truck insulated or picked up with fish stored in Styrofoam filled with ice. The low temperature of the fish needs to be maintained during the sale process by sprinkling ice on the fish. Fish received need to be washed with clean water before being sold. The fish thawing process must be carried out according to standards. During fish handling, wear gloves and clean equipment to avoid bacterial contamination. The unsold fish were stored in the refrigerator or at a chilling temperature. Applying good hygiene can reduce bacterial contamination from the environment to the fish surface. Keeping the cold chain during the distribution process can inhibit the activity and growth of bacteria.







