Background
Microalgae are widely used as bioreactors to produce heterologous proteins with high added value because they provide a simple mass culture system. They are a natural system in which to produce various useful materials (Walker et al. 2005). In some types of microalgae, the oil content exceeds 80 % of the dry weight of the algal biomass, which is a potentially useful bioenergy source (Banerjee et al. 2002). Recently, many studies have been conducted in the fields of genetic and metabolic engineering to improve the economic production of microalgal diesel (Chisti 2008; Mitra and Melis 2008).
Dunaliella salina belongs to the phylum Chlorophyta in the order Volvocales. It is a photosynthetic, unicellular motile biflagellate microalga that can be cultured simply, rapidly, and inexpensively (Hosseini Tafreshi and Shariati 2009). D. salina is also extremely adaptable to various environments and can live at various salt concentrations, ranging from 0.05 to 5.0 M sodium chloride (Feng et al. 2014). It can accumulate useful materials such as carotenoids, lipids, vitamins, and minerals, and particularly β-carotene, which is a source of pigments, antioxidants, and anticancer agents for use in foods, cosmetics, and the pharmaceutical industry (Tan et al. 2005; Hosseini Tafreshi and Shariati 2009). D. salina is a natural protoplast, lacking a rigid cell wall, and exogenous genes can easily be introduced into the cells with molecular techniques. Therefore, D. salina is considered a good bioreactor for the production of heterologous proteins, including recombinant proteins (Feng et al. 2014).
The stable genetic transformation of D. salina has been achieved in studies that predominantly focused on selectable markers, promoters, and procedures for the efficient transformation of D. salina cells with exogenous DNA (Wang et al. 2007). However, the availability of a reliable transformant selection protocol is a prerequisite for the development of a practical transformation system (Jiang et al. 2005). Most transgenic microalgal selection involves either (1) selection based on antibiotic or herbicide resistance or (2) selection based on the complementation of metabolic or photosynthetic mutants (Potvin and Zhang, 2010). The first method is predominantly used in microalgal transformation (Coil 2006; Potvin and Zhang, 2010). However, an unstable phenotype has been reported when an antibiotic resistance gene was used as a selectable marker in the microalgal transformation system (Cerutti et al. 1997; Jiang et al. 2005). Moreover, D. salina is naturally resistant to many of the antibiotics commonly used for the selection of genetically transformed plant cells, such as spectinomycin, kanamycin, hygromycin, and G418 (Jiang et al. 2005; Akbari et al. 2014). To overcome this limitation, an effective selectable marker for D. salina is required.
The objective of this study was to develop an effective selectable marker with which to establish a transformation system for D. salina. To do so, the sensitivity of D. salina to chloramphenicol and Basta® (Bayer CropScience, Monheim, Germany) was tested and the feasibility of using this sensitivity as a selectable marker in the production of transgenic D. salina was investigated.
Methods
D. salina (KMMCC-1064) was obtained from the Korea Marine Microalgae Culture Center (KMMCC). The cells were cultured at 24 °C under 4500–5000 lux illumination with a 14-h light/10-h dark cycle in f/2 medium (Guillard and Ryther 1962). Cells in logarithmic phase were used for the experiments.
PCR was used to amplify the ITS regions of D. salina with the primer pair designed by Li et al. (2007), based on conserved sequences in 18S and 28S rDNA: ITSF (5′-GGAAGGAGAAGTCGTAACAAGG-3′) and ITSR (5′-TCCTCCCTTATTGATATGC-3′). The PCR cycling parameters were as follows: 94 °C for 4 min; 35 cycles of 94 °C for 1 min, 50 °C for 45 s, and 72 °C for 2 min; and a final extension at 72 °C for 10 min. The PCR product was sequenced directly on an ABI 3100 DNA Analyzer (Applied Biosystems, USA) with the same primers.
To determine the phylogenetic relationships of D. salina, the ITS region sequence was aligned with the corresponding sequences of Dunaliella strain sequences obtained from the GenBank database, using ClustalX (Thompson et al. 1999) in BioEdit Ver 7.0.9.0 (Hallmann and Rappel 1999). A neighbor-joining (NJ) analysis was performed with Kimura’s two-parameter model in PAUP 4.10b (Swofford 2002). Bootstrap support values were derived from 1000 randomized replicate datasets. Statistical reliability was evaluated based on the posterior probability obtained from a 50 % majority-rule consensus tree. Five species of Chlamydomonas were used as the outgroup.
The selectable agents chloramphenicol and Basta® were examined for their ability to prevent the growth of wild-type D. salina cells in liquid culture. The final concentration of chloramphenicol (Sigma, USA) was 0–1500 μg/ml and that of Basta® (Bayer CropScience, Germany) was 0–40 μg/ml. The number of cells was counted with a hemocytometer under a light microscope every 1–2 days. The initial cell density of D. salina was 1.0 × 106 cells/ml and each experiment was performed separately three times.
The pat cassette, the expression vector used in this study, containing a pat gene under the control of Cauliflower mosaic virus 35S promoter (p35S) and 35S terminator (t35S), was amplified by PCR from the genomic DNA of genetically modified corn with the primers HindIII pat cassette 2F (5′-ATAAAGCTTTCCCATGGAGTCAAAGATTC-3′) and XhoI pat cassette 1R (5′-ATACTCGAGCAGGTCGACTCTAGAGGAT-3′). The PCR cycling parameters were as follows: 94 °C for 2 min; 33 cycles at 94 °C for 30 s, 58 °C for 30 s, and 72 °C for 1 min; and a final extension at 72 °C for 7 min. The PCR products were purified with the AccuPrep Gel Purification Kit (Bioneer, Korea), according to the recommendations of the manufacturer, and then transferred into the pGEM-T Easy Vector System (Promega, Madison, WI, USA) with TA cloning. The configuration and sequence of the expression vector was confirmed by sequencing the region from the 5′ end of p35S to the 3′ end of t35S (Fig. 1).
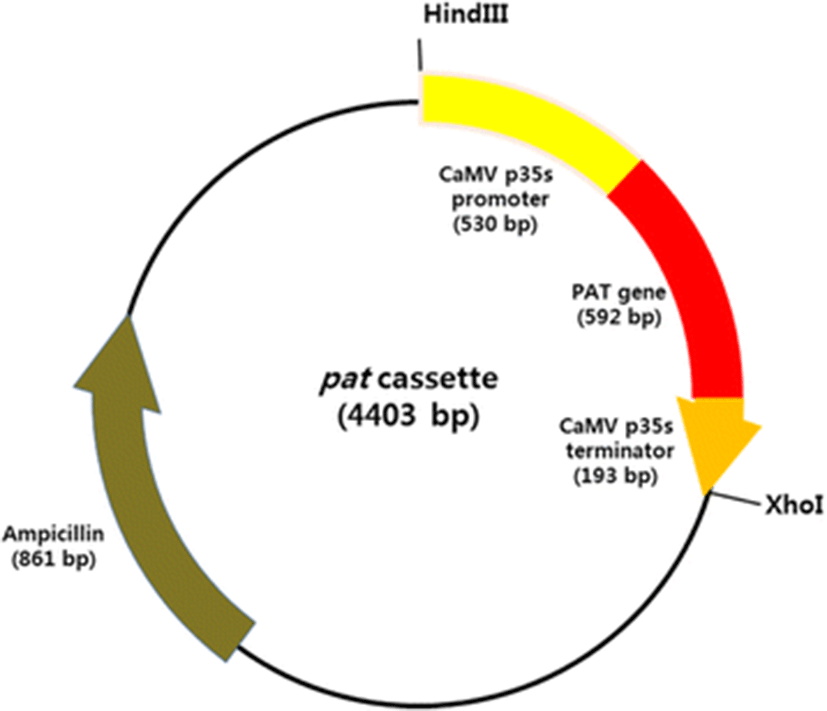
The transformation for D. salina was conducted with the particle bombardment method using the Biolistic PDS-1000/He Particle Delivery System (Bio-Rad, USA). Gold microcarriers (0.6 μm in diameter) were coated with the pat cassette (Fig. 1), according to the methodology of Sanford et al. (1993).
Wild-type D. salina cells grown to logarithmic phase were harvested by centrifugation and resuspended in f/2 medium. The resuspended cells (final cell density 1 × 107 cells/ml) were spread on 1.5 % agar plates containing 0.5 M NaCl and bombarded with the plasmid-coated microcarriers under helium pressure of 900 psi from a distance of 3 cm. The bombarded cells were immediately collected and cultured in liquid f/2 medium at 25 °C for 48 h in the dark.
To select the transformed cells, 5 μg/ml Basta® was added to the suspension of bombarded cells. After 48 h, the surviving cells in the liquid selection medium containing Basta® were spread on solid f/2 medium containing 1.5 % agar.
The genomic DNA for PCR analysis was extracted from the cells surviving in f/2 liquid medium containing 5 μg/ml Basta®. PCR was used to detect the integration of the pat gene into D. salina with primers P35sF (5′-ATTGATGTGATATCTCCACT-3′) and T25R (5′-TGAGCGAAACCCTATAAGAA-3′), designed based on the sequence of the pat gene. The PCR cycling parameters were as follows: 94 °C for 2 min; 33 cycles at 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 1 min; and a final extension at 72 °C for 7 min. The PCR products were evaluated by electrophoresis on 1.5 % agarose gels stained with ethidium bromide.
Results
An ITS gene fragment of 600 bp was amplified from the genomic DNA of D. salina. A BLAST search of the ITS region sequence showed that ITS1, 5.8S rDNA, and ITS2 of the D. salina strain used in this study showed 99 % sequence identity with D. salina and Dunaliella sp.(HQ590542.1) (Fig. 2a).
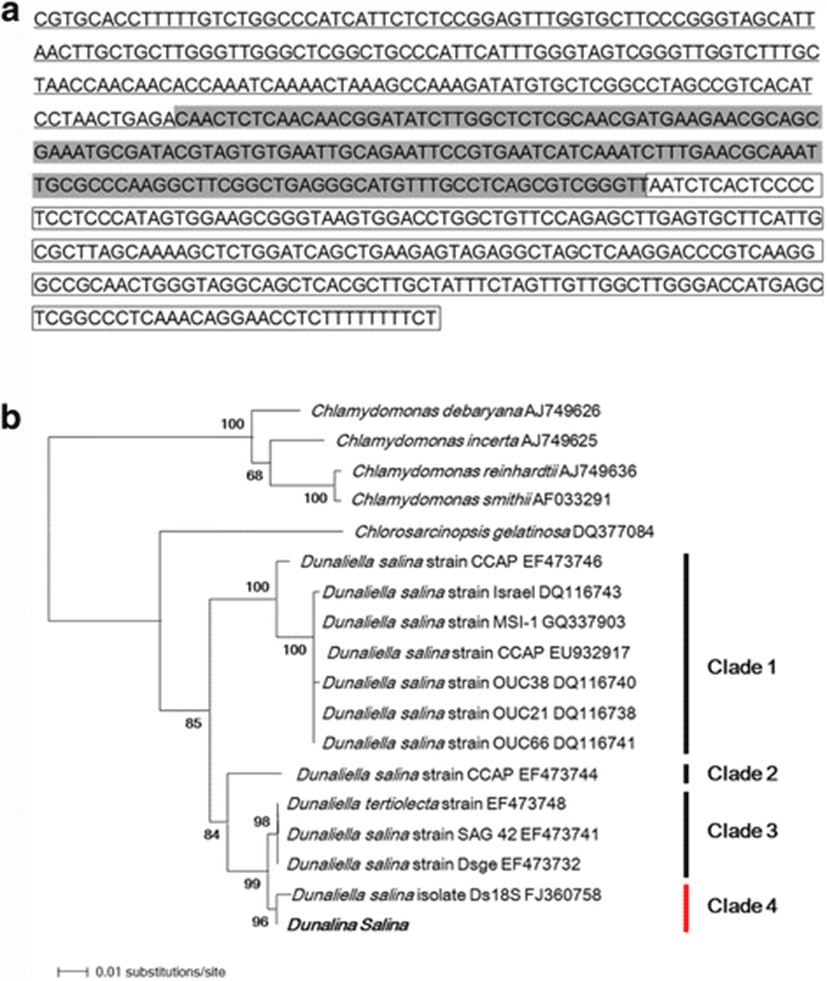
A phylogenetic tree was constructed with the neighbor-joining method to determine the phylogenetic relationships between several species of the genus Dunaliella based on ITS region sequences (Fig. 2b). The D. salina strain used in this study had different genetic relationships with the four species of the genus Chlamydomonas used as the outgroup. The species of the genus Dunaliella clustered into four clades (85 % bootstrap support), designated clades 1, 2, 3, and 4, as shown in Fig. 2b. The D. salina strain used in this study belonged to clade 4 (96 % bootstrap support) and shared 99 % sequence identity with D. salina isolate Ds18S (GenBank accession number FJ360758). These findings confirm that the cells used in this study were a D. salina strain.
The result of the sensitivity test to chloramphenicol showed that the number of D. salina cells decreased gradually from 1.0 × 106 cells/ml to 6.0 ± 2.0 × 105 cells/ml and 3.67 ± 0.6 × 105 cells/ml in 1000 and 1500 μg/ml chloramphenicol, respectively, on day 2, and that 1000 and 1500 μg/ml chloramphenicol completely inhibited the growth of D. salina in 8 and 6 days, respectively (Fig. 3a).
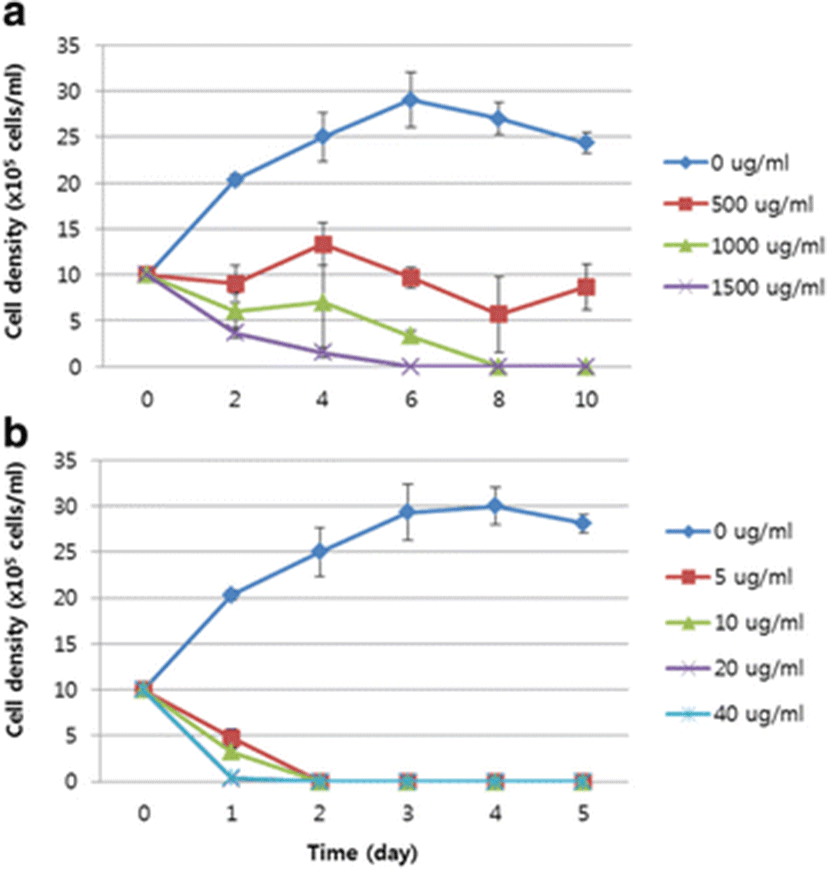
In 5, 10, 20, and 40 μg/ml Basta®, the number of cells was reduced rapidly (in 1 day) to 4.83 ± 1.1 × 105 cells/ml, 3.57 ± 0.4 × 105 cells/ml, 0.27 ± 0.25 × 105 cells/ml, and 0.33 ± 0.35 × 105 cells/ml, respectively. The D. salina cells died completely in 2 days at all treatment concentrations (Fig. 3b). These findings suggest that D. salina is more sensitive to Basta® than to chloramphenicol, and that Basta® is a more effective selectable agent for D. salina than chloramphenicol.
To screen the D. salina cells transformed by bombardment with the pat cassette plasmid were cultured first in liquid f/2 medium containing 5 μg/ml Basta® and then on f/2 agar medium. Colonies of bombarded D. salina appeared on the agar plate, whereas no colonies of the control wild-type D. salina appeared (Fig. 4a).
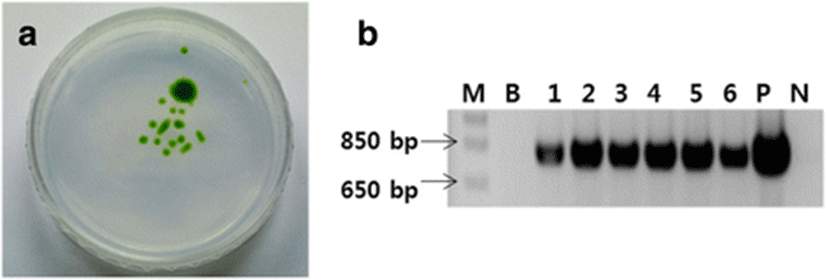
A PCR analysis was performed using the genomic DNA of individual colonies of bombarded D. salina picked from the plate. A fragment of ~796 bp was amplified from the DNA of all bombarded colonies but not from the DNA of wild-type D. salina (Fig. 4b). These findings indicate that the pat gene was successfully inserted into the D. salina.
Discussion
In this study, we aimed to develop an effective selectable marker to establish a transformation system for D. salina, a very valuable bioreactor organism.
The morphologies of individual species of Dunaliella vary considerably at different stages of their growth and development, and under different environmental conditions. Therefore, possible errors in taxonomic assignments have been reported when they are only based on morphological analyses (Borowitzka and Siva, 2007; Tran et al. 2013). The ITS region of the 18S rRNA gene and the large subunit of the ribulose bisphosphate carboxylase gene (rbcL) are widely used as effective molecular tools in microalgal characterization and biodiversity studies (Preetha et al., 2012). Of these molecular markers, the ITS region is long, with much sequence variation, so sequence comparisons of the ITS region are used to determine the genetic relatedness and phylogenic and taxonomic status of Dunaliella (Han et al., 2009; Hejazi et al., 2010). Sequencing and molecular phylogenetic analyses of the ITS region confirmed the identity of our D. salina strain, which clustered in a monophyletic group on the phylogenic tree and showed 99 % sequence similarity with D. salina isolate Ds18S (GenBank accession number FJ360758).
The selection of transformants is an integral and crucial part of any microalgal transformation system (Tan et al. 2005). A selectable marker gene allows the growth of transformed cells in the presence of the corresponding selectable agent, such as an antibiotic or herbicide, which is toxic to untransformed cells (Bohorova, 1999). Numerous previous studies have used selectable marker genes such as the cat gene, encoding chloramphenicol resistance, or the pat gene, encoding resistance to the herbicide phosphinothricin, which can be used to select transformed Dunaliella cells (Jiang et al. 2005; Akbari et al. 2014).
Chloramphenicol, also called chloromycetin, inhibits the peptidyl transferase activity of bacteria. Chloramphenicol acetyl transferase, encoded by the cat gene, detoxifies chloramphenicol by acetylating it (Ziemienowicz 2001). D. salina is naturally resistant to various antibiotics, including streptomycin, kanamycin, hygromycin, spectinomycin, and G418, whereas chloramphenicol has been reported to inhibit the growth of D. salina (Tan et al. 2005). In this study, D. salina cells (1.0 × 106 cells/ml) died in 8 and 6 days when treated with 1000 and 1500 μg/ml chloramphenicol, respectively. Therefore, the concentration of chloramphenicol used to select transgenic D. salina within 7 days was >1000 μg/ml (Fig. 3a). However, the concentration observed in our study was much lower than that observed in several other studies. Geng et al. (2003) reported that D. salina died in 3 weeks in 60 μg/ml chloramphenicol, and Tan et al. (2005) reported that D. salina died in 400 μg/ml chloramphenicol. These discrepancies are attributed to differences in the cell densities used and the periods of antibiotic treatment.
Basta® is a herbicide containing glufosinate ammonium (l-phosphinothricin), which causes the death of microalgae by blocking the activity of an enzyme involved in the synthesis of the amino acid glutamine (Hoerlein 1994; Kang et al., 2014). A test of the sensitivity of D. salina to Basta® showed that a concentration of 5 μg/ml was sufficient to completely inhibit the growth of D. salina within 2 days (Fig. 3b). Various other studies have reported that D. salina cells are extremely sensitive to Basta®. Jiang et al. (2005) reported that the concentrations inhibiting the growth of D. salina were 0.5 μg/ml in liquid medium and 1 μg/ml in solid medium, and Tan et al. (2005) reported that a concentration of 20 μg/ml was sufficient to completely inhibit the growth of D. salina.
Our findings suggest that the appropriate concentrations of chloramphenicol and Basta® for the selection of transgenic D. salina are 1500 and 5 μg/ml, respectively, and that D. salina cells are more sensitive to Basta® than to chloramphenicol. Therefore, Basta® may be an efficient selectable agent for use in the genetic transformation of D. salina. To evaluate the utility of the pat gene as a selectable marker gene for transgenic D. salina, a plasmid containing the pat gene was introduced into D. salina with a particle bombardment system. The Basta®-resistant cells were checked with a molecular analysis. PCR indicated that the pat cassette was inserted into the D. salina cells (Fig. 4). The findings of this study suggest that the pat gene can be used as a selectable marker for transgenic D. salina to produce useful recombinant proteins.
Conclusions
We concluded that D. salina cells are more sensitive to Basta® than to chloramphenicol. Therefore, Basta® may be an efficient selectable agent for use in the genetic transformation of D. salina. Also, as a result of the possibility of using the pat gene as a selectable marker gene, we concluded that the pat gene can be used as a selectable marker for transgenic D. salina to produce useful recombinant proteins.