Introduction
Pleuronectidae, commonly known as the flatfish family, comprises 40 genera and 103 species worldwide. Japan has 17 genera and 33 species, whereas South Korea has 18 genera and 25 species reported (Lee et al., 2019). Among the 25 species reported in South Korea, 13, including the yellowfin sole, have either limited records in the literature or very low occurrence frequencies. The remaining 12 species, Pleuronectes yokohamae, Pleuronectes herzensteini, Hippoglossoides pinetorum, Hippoglossoides dubius, Kareius bicoloratus, Pleuronichthys cornutus, Clidoderma asperrimum, Eopsetta grigorjewi, Glyptocephalus stelleri, Platichthys stellatus, Tanakius kitaharae (=Glyptocephalus kitaharae), and Microstomus achne are considered important edible fishery resources. However, these 12 species are difficult to classify morphologically due to their similar appearances. Except for the Greenland halibut, the catch quantities are not recorded by species but grouped under the generic term “flatfish.”
Recently, various seafood products have been imported and distributed throughout South Korea. According to the Fishery Information Portal System (www.fips.go.kr) of the Ministry of Oceans and Fisheries, the import volume of flatfish has more than doubled since 2017, reaching over 3,500 tons. Owing to the wide variety of species and their similar appearance, flatfish are susceptible to economic fraud, where cheaper species are misrepresented and sold as more expensive ones, leading to frequent occurrences of economically motivated adulteration in the domestic seafood market. Given the high potential for disputes regarding the identification of species and their origins in the flatfish family, further research is required to obtain genetic information through genetic characteristic analyses and establish a reliable classification system.
Previous studies on the flatfish family have mainly focused on the morphological characteristics and distribution of species, such as Verasper variegatus, P. cornutus, P. yokohamae, G. stelleri, and T. kitaharae. Additionally, there have been molecular phylogenetic studies on the Korean flatfish family. However, methods of species identification of the flatfish family using genetic analysis remains unreported.
The species classification of aquatic organisms serves as a fundamental unit for the management of biological resources. There is a growing awareness of the importance of species classification, given its importance in ecosystem research to ensure food safety through the prevention of misrepresentation of species, and combat counterfeit and substandard food products.
Among the various methods currently used for species identification, DNA-based species identification techniques that utilize molecular biology approaches offer higher reliability in analyzing processed samples, including those subjected to high temperatures and drying during the manufacturing process, owing to the stability of their physicochemical compositions (Axayácatl & Juan, 2008). Among these methods, the multiplex species-specific polymerase chain reaction (MSS-PCR) analysis allows for species identification by only performing PCR amplification followed by electrophoretic analysis without the need for pretreatments with restriction enzymes. MSS-PCR is simpler and exhibits higher reproducibility compared to methods such as restriction fragment length polymorphism (RFLP) and amplified fragment length polymorphism (Acar et al., 2017; Kang et al., 2015; Kim et al., 2014; Noh et al., 2017; Rasmussen & Morrissey, 2008). Furthermore, by analyzing intra-and interspecies polymorphisms using nucleotide sequences and designing primers, it is possible to mitigate false-positive and false-negative results resulting from differences in genetic variation (Axayácatl & Juan, 2008; Hsieh & Hwang, 2004; Hsieh et al., 2010).
In this study, we focused on a variety of flounder species within the family Pleuronectidae that are challenging to differentiate owing to their similar morphologies. Specifically, we analyzed the genetic characteristics of 11 species from 10 genera: G. stelleri, P. herzensteini, T. kitaharae, C. asperrimum, K. bicoloratus, M. achne, P. yokohamae, E. grigorjewi, P. stellatus, Cleisthenes pinetorum, and Limanda aspera. Through genetic analysis, we gained insights into their genetic characteristics. We analyzed intra-and interspecies genetic variations using the cytochrome c oxidase subunit I (COI) gene region of mitochondrial DNA (mtDNA), which has been proposed as the species identification barcode region by the Consortium for the Barcode of Life. Additionally, we aimed to establish MSS-PCR conditions, enabling the identification of 11 flounder species from the Pleuronectidae family in a single PCR reaction by exploring species-specific positions with distinct inter-species variations (Ward et al., 2009).
Our findings will contribute to the systematic resource management and food safety of imported and domestic flounder species within the Pleuronectidae family by establishing an MSS-PCR analysis method using DNA-based molecular biology techniques.
Materials and Methods
Overall, 322 samples from 11 species belonging to 10 genera of flounder were obtained from G. stelleri, P. herzensteini, T. kitaharae, C. asperrimum, K. bicoloratus, M. achne, P. yokohamae, E. grigorjewi, P. stellatus, C. pinetorum, and L. aspera (Table 1). These samples were procured from domestic fish markets between January and June 2019 or were provided by the Marine and Fishery Resource Donation and Preservation Institute at Pukyong National University. Of the 322 samples, 77 were used for primer design, and the remaining 245 for primer validation.
DNA was extracted from samples using a DNeasy® Blood & Tissue Kit (Qiagen GmbH, Hilden, Germany). Approximately 20 mg of tissue from each sample was used for genomic DNA (gDNA) extraction, following the instructions provided in the manual accompanying the DNeasy® Blood & Tissue Kit. The extracted gDNA was quantified using a Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and stored at –20°C until further genetic analysis.
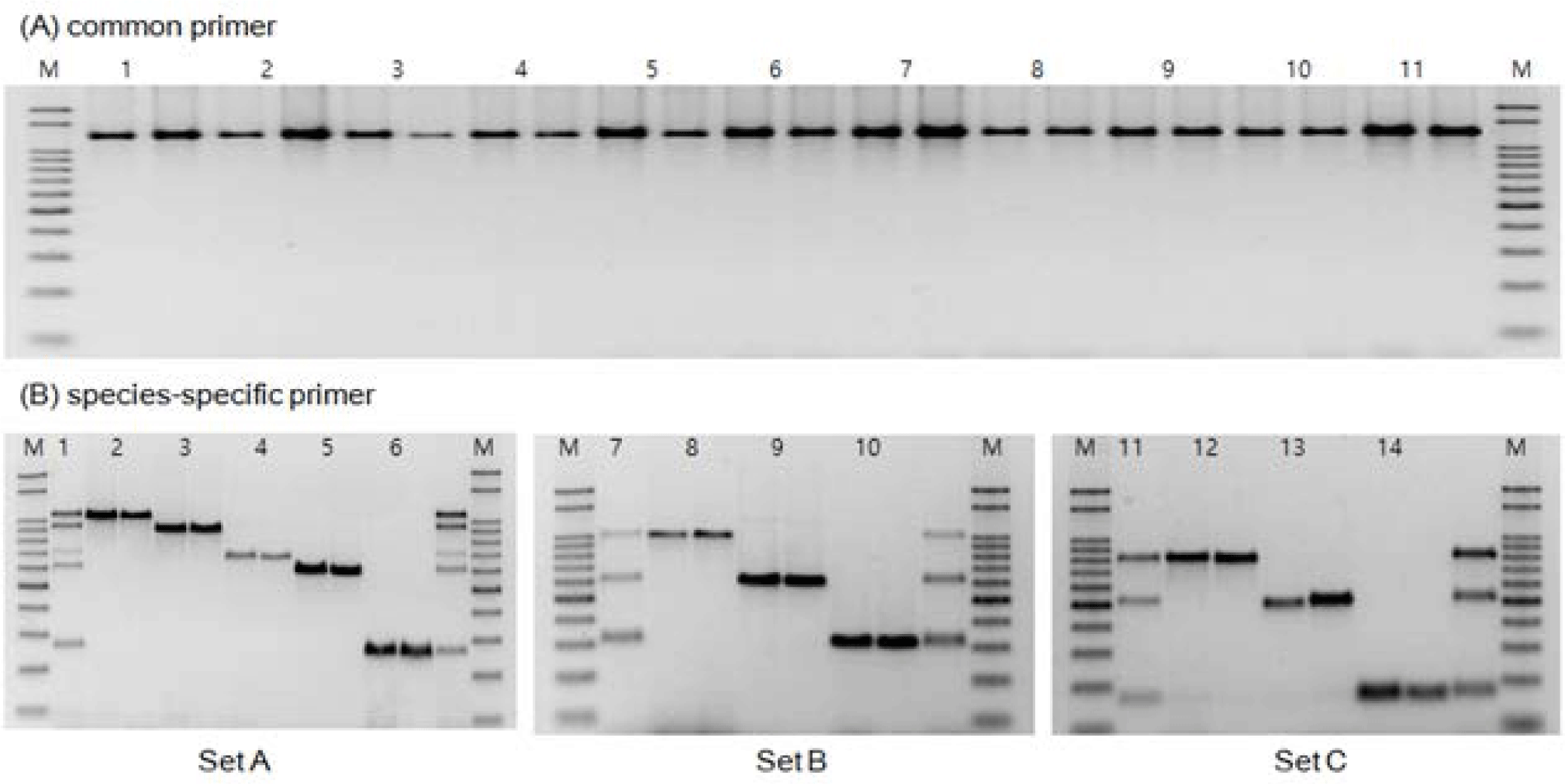
To explore inter- and intra-species variations and identify conserved regions within the COI gene region of mtDNA, we aligned mtDNA nucleotide sequences from 11 flounder species (G. stelleri, MH032428.1; P. herzensteini, MH032530.1; Glyptocephalus kitaharae, MH032424.1; C. asperrimum, MH032407.1; K. bicoloratus, MH032487.1; M. achne, MH032470.1; P. yokohamae, KT878309.1; E. grigorjewi, MH032414.1; P. stellatus, EF424428.1; C. pinetorum, MH032404.1; L. aspera, MH032456.1) registered in the National Center for Biotechnology Information (NCBI) GenBank. We used BioEdit (http://mbio. ncsu. edu/BioEdit/biodeit.html) to align the nucleotide sequences. Based on the conserved regions identified in the alignment, we designed a specific forward primer (GJM-F128) and reverse primer (GJM-R1370) to amplify the COI gene region of these 11 species (Fig. 1 and Table 2).
| Oligo name | Sequence (5’ → 3’) | Mer | Tm (°C) | GC (%) | Product size |
|---|---|---|---|---|---|
| GJM-F128 | GCA CAC GCC TTT GTA ATA AT | 20 | 59 | 40 | 1,242 bp |
| GJM-R1370 | CGT TTG GCT GTA AAT GCT TC | 20 | 62 | 45 |
To amplify of the COI gene region, the following reaction mixture was prepared:1 μL of extracted gDNA, 2 μL of 10 × Taq buffer, 0.4 μL of 2.5 mM dNTP mixture, 0.8 μL of 10 pmol forward primer, 0.8 μL of 10 pmol reverse primer, and 0.5 units of HS-Taq polymerase (Anti-HS Taq, TNT Research, Jeonju, Korea), making a total volume of 20 μL. The PCR conditions were set using ABI Verity Fast Thermal Cyclers (Applied Biosystems, Foster, CA, USA), with an initial pre-denaturation step at 95°C for 10 min, followed by denaturation at 95°C for 40 s, annealing at 54°C for 40 s, and extension at 72°C for 50 s, repeated for 35 cycles. The final extension step was performed at 72°C for 7 min. The amplified PCR products were analyzed using an automated DNA sequencing instrument with an ABI 3730XL DNA analyzer (Applied Biosystems).
The bidirectional nucleotide sequences of the COI gene region from 11 species of the Pleuronectidae family were aligned using the DNASTAR SeqMan software (DNASTAR, Madison, WI, USA). Finally, a 1,262 bp sequence of the COI gene region was obtained. The obtained sequences were analyzed to identify species-specific single nucleotide polymorphisms (SNPs), excluding intraspecific variations caused by point mutations. Species-specific primers were designed to differentiate each species based on the size differences of the PCR amplification products during gel electrophoresis (Fig. 2). Primers were designed based on the size of the PCR amplification products for each species to achieve species discrimination. Primers were designed with a species-specific SNP located at the 3’ end to enable the amplification of only the target species. Eleven forward primers were used in this study. Species-specific forward primers were also designed to allow amplification at similar melting temperature (Tm) values for convenience in the experimental procedures. For five species of Pleuronectidae, G. stelleri, T. kitaharae, C. asperrimum, E. grigorjewi, and C. pinetorum, the primers were designed at 58°C, while for the six species, P. herzensteini, M. achne, P. stellatus, K. bicoloratus, Pleuronectes yokohamae and L. aspera the primers were designed at 62°C. We also considered factors such as GC content to prevent self-dimer formation while designing the primers (Table 3).
For MSS-PCR amplification, three sets of primer mixtures capable of distinguishing 11 Pleuronectidae species were prepared. The first set comprised five species: G. stelleri, T. kitaharae, C. asperrimum, E. grigorjewi, and C. pinetorum. The second set comprised three species, P. herzensteini, M. achne and P. stellatus. The third set comprised three species: K. bicoloratus, P. hokohamae, and L. aspera.
The reaction mixture for MSS-PCR amplification was composed of 1 μL of gDNA extracted from each sample, 1 μL of 10x Ex Taq buffer, 0.2 μL (for 5 species) to 0.3 μL (for 3 species) of a 2.5 mM dNTP mixture, 0.3 μL (for 3 species) to 0.4 μL (for 5 species) of 10 pmol forward primer, 0.6 μL (for 3 species) to 0.8 μL (for 5 species) of 10 pmol reverse primer, and 0.5 units of HS-Taq polymerase (Anti-HS Taq) in a total volume of 10 μL.
The PCR conditions were as follows: pre-denaturation at 95°C for 10 min, denaturation at 95°C for 40 s, annealing at 58°C (for 5 species) to 62°C (for 3 species) for 40 s, and extension at 72°C for 50 s. These cycles were repeated 30 times, followed by a final extension at 72°C for 7 min. Amplification was performed using the ABI Verity Fast Thermal Cycler.
PCR was performed using an ABI Verity Fast Thermal Cycler under the same conditions described above, with different annealing temperatures for each set. Subsequently, the amplified PCR products were separated on a 2% agarose gel at 100 V for 40 min by gel electrophoresis. The gel was stained with 1X loading dye (Dynebio, Seongnam, Korea), and the PCR products were visualized using a Gel Doc image analysis system (ATTO, Tokyo, Japan).
Results
Based on the COI gene nucleotide sequences registered in the NCBI GenBank database, we explored conserved regions with minimal variation and designed species-specific forward and reverse primers for 11 species of the Pleuronectidae family (Fig. 1). The designed primers were applicable to all 11 Pleuronectidae species. They were used as reference markers to confirm amplification by PCR and served as a universal primer set for the Pleuronectidae family.
The COI gene region (1,262 bp) obtained from the 11 species was analyzed using DNA Sequence Polymorphism software (DnaSP, ver. 5.10.01) with universal primers designed to apply to all 11 species. In the analyzed sequences, excluding intraspecies genetic variations, distinct interspecies variations were observed in the form of species-specific SNPs across all 11 species (Fig. 1). Based on these observations, species-specific forward primers were designed with SNPs located at the 3´ end, allowing for differentiation among the 11 species of the Pleuronectidae family with sequence differences of approximately 100 bp or more (Fig. 1 and Table 3).
Haplotype diversity analysis revealed the following values: C. pinetorum 0.855, P. herzensteini 0.389, P. yokohamae 0.896, C. asperrimum 0.873, K. bicoloratus 0.495, G. stelleri 0.925, M. achne 0.889, T. kitaharae 0.923, E. grigorjewi 0.795, and L. aspera 0.948. However, P. stellatus exhibited only one haplotype, indicating an absence of genetic diversity.
To determine the maximum limit of the annealing temperature, gradient PCR reactions were performed using conditions ranging from 58°C to 62°C. PCR amplification was performed using forward primers specific to each species and a universal reverse primer (GJM_1370-R) specific to the Pleuronectidae family. The amplified products were visualized through electrophoresis on a 1.8% agarose gel. The optimal temperature for each primer was determined based on the temperature at which the highest species-specific amplification was observed (Table 3).
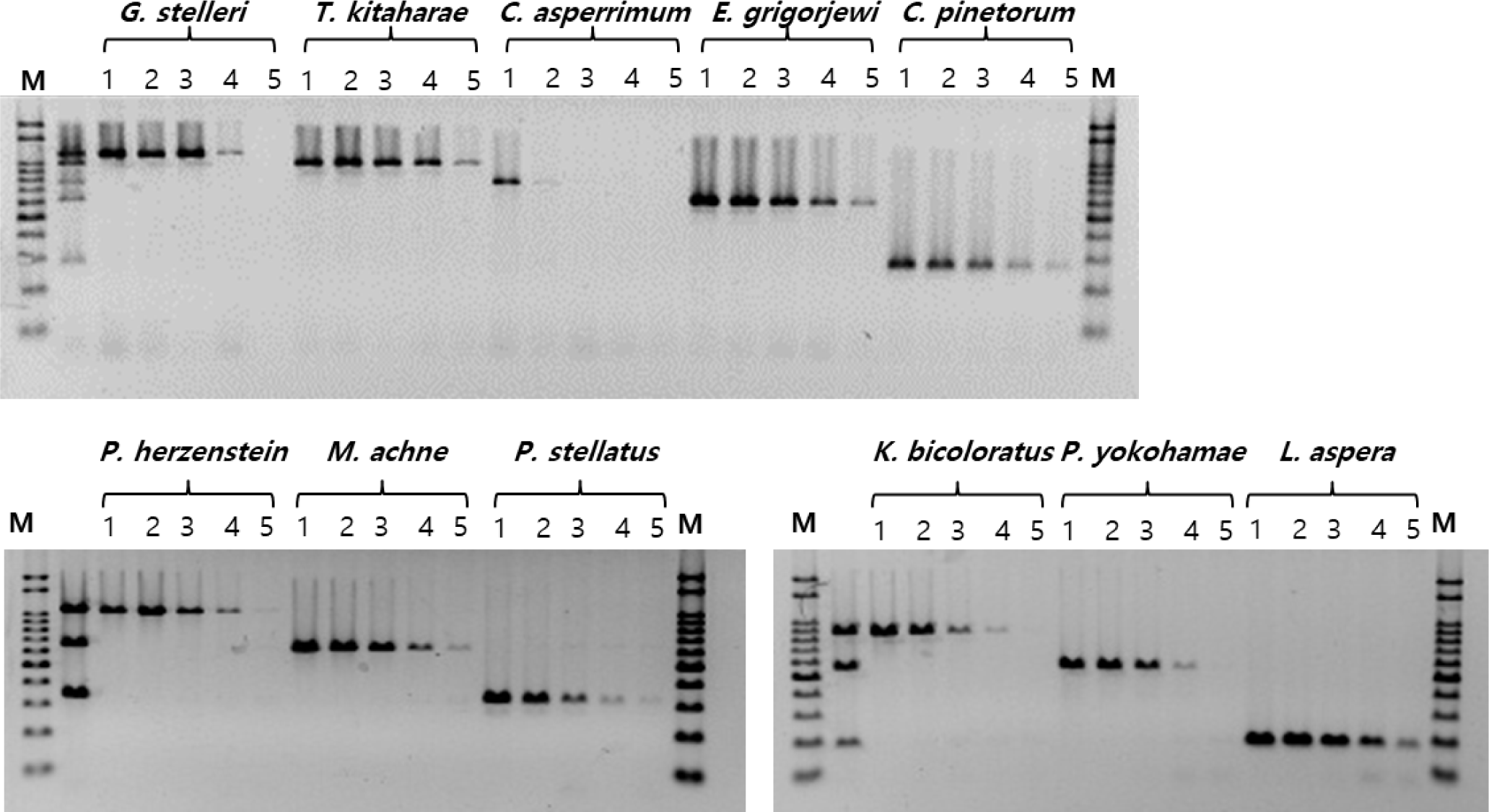
Upon verification of the MSS-PCR products containing equal amounts of forward primers by agarose gel electrophoresis, clear DNA amplification was observed for each species. The amplified products were of the expected size for each species: G. stelleri (1,064 bp), T. kitaharae (933 bp), C. asperrimum (709 bp), P. hokohamae (571 bp), C. pinetorum (259 bp), P. herzensteini (1,006 bp), M. achne (613 bp), P. stellatus (312 bp), K. bicoloratus (815 bp), P. hokohamae (517 bp), and L. aspera (159 bp). No primer dimers or nonspecific amplification was observed. Furthermore, species-specific PCR amplification was confirmed, without cross-reactivity in the DNA mixtures of each primer set (Fig. 3).
To assess the sensitivity of the MSS-PCR analysis, DNA from 11 species of the Pleuronectidae family was quantified at concentrations of 50, 10, 1 ng/μL, 0.1 ng/μL, and 0.01 ng/μL for experimentation. The PCR analysis sensitivity for each species was as follows: C. asperrimum exhibited detection at 50 ng/μL, while G. stelleri, P. herzensteini, P. stellatus, K. bicoloratus, and P. hokohamae showed detection down to 0.1 ng/μL. T. kitaharae, E. grigorjewi, C. pinetorum, M. achne, and L. aspera demonstrated detection capabilities at 0.01 ng/μL of DNA concentration (Fig. 2).
Discussion
The diverse and morphologically similar nature of fish species within the family Pleuronectidae makes species identification based on morphological characteristics challenging. However, the application of molecular biology techniques, such as genetic analysis, has facilitated the identification of clear species (Kwun & Kim, 2010, 2016). mtDNA analysis for biological species identification offers advantages over nuclear DNA analysis because mtDNA has a higher copy number, which enables the extraction of a significant amount of DNA, even with limited sample quantities (Cohen et al., 2009; Matsui et al., 2012). Furthermore, mtDNA is maternally inherited and possesses only one allele, thereby avoiding the ambiguity of genetic sequences caused by allele heterozygosity. Due to its relatively higher mutation rate than nuclear DNA, mtDNA is commonly used in species identification and molecular phylogenetic studies involving closely related species (Matsui et al., 2012). However, the high mutation rate of mtDNA can also lead to intraspecies genetic variation, making it important to validate the accuracy of analytical methods by analyzing multiple samples in studies focusing on species identification among closely related species.
The MSS-PCR analysis method offers advantages over traditional non-specific primer-based methods, such as RFLP and RAPD, in terms of reproducibility, time, and cost; most importantly, it provides clear and repeatable results for species discrimination (Kim & Lee, 1990; Kim et al., 2009; Lin & Hwang, 2008; Sezaki et al., 2005). Recently, MSS-PCR analysis has been applied to the development of species analysis techniques for various aquatic products, including shrimp paste, smoked eel, mackerel, and horse mackerel (Kim et al., 2009, 2014; Noh et al., 2016). In MSS-PCR analysis, it is crucial to design primers with size differences in the PCR products to ensure clear species discrimination and establish conditions for multiplex PCR that prevent non-specific PCR reactions and maintain appropriate annealing temperatures (Koh et al., 2011; Silfvergrip, 2009).
In this study, we developed and established conditions for the MSS-PCR analysis method, which allows multiplex PCR targeting of 11 species of the Pleuronectidae family where morphological species identification is challenging. We analyzed the COI gene region of mtDNA and validated the applicability of the primers for species specificity, reproducibility, and efficiency in 11 species.
This analytical method enables accurate species identification of processed flatfish products or imported seafood, where traditional morphological identification is difficult. This can contribute to the establishment of a sound distribution system and ensure food safety. Therefore, they can be effectively utilized to enhance the safety and reliability of seafood products.
