Background
The aquatic culture of the olive flounder (Paralichthys olivaceus) has been widespread in Korea. The farming of juvenile olive flounder, however, has caused a lot of problems due to the occurrence of various diseases (Ototake and Matsusato 1986; Park 2009). The juvenile flounder is difficult to manage and is weak against diseases, and the mortality rates have been economically damaging (Jee et al. 2001).
Small GTPases are well known as one of the signal transduction factors of immune systems (Narumiya 1996; Scheele et al. 2007). Some documents indicated that small GTPases are related to virus infection in the shrimp (Wu et al. 2007; Liu et al. 2009; Zhang et al. 2010). Also, the small GTPases of zebra fish have provided a firm basis of an innate immune system in vertebrates (Salas-Vidal et al. 2005). The authors therefore studied on ADP-ribosylation factor, which is a member of the GTP-binding proteins, from the olive flounder to investigate the relation between cytoskeleton remodeling and the olive flounder immune system.
The ADP-ribosylation factor (ARF) proteins are small GTP-binding proteins, and they are involved in membrane dynamics and the regulation of actin cytoskeleton organization (D’Souza-Schorey and Chavrier 2006; Myers and Casanova 2008). The ARF can be classified into three groups based on the peptide sequence, protein molecular weight, gene structure, and phylogenetic analysis, as follows: class I including ARF1, ARF2, and ARF3; class II including ARF4 and ARF5; and class III including only ARF6 (Myers and Casanova 2008; Tsuchiya et al. 1991). The class I and class II ARFs are mainly associated with the Golgi complex, although they also function in endosomal compartments (Myers and Casanova 2008). In addition, ARF proteins were identified as activators of phospholipase D (PLD) (Luo et al. 1998). ARF1 was shown to recruit coat proteins to the Golgi membranes when it is bound to GTP (Balch et al. 1992). The hydrolysis and binding of GTP by ARF1 were originally linked to the assembly and disassembly of vesicle coats (Nie and Randazzo 2006).
The radiation of teleosts has been attributed to a genome-DNA event during the evolution of the teleosts (Venkatesh 2003). Although a number of ARFs, from those of micro-organisms to those of mammals, have been studied, a lack of studies on the duplicated ARF genes in the olive flounder persists. The authors therefore isolated and characterized one of the class I duplicated ARF genes.
Methods
Total RNA was extracted by using GeneAll® Hybrid-R™ Total RNA (GeneAll Biotechnology Co., Ltd., Korea) following the manufacturer’s instructions from 12 tissues, including the brain, eye, gullet, heart, liver, stomach, muscle, kidney, spleen, pyloric ceca, intestine, and gill tissues, of healthy Paralichthys olivaceus. And then, we carried out 5′- and 3′-rapid amplification of cDNA ends (RACE) by using SMART™ RACE cDNA Amplification Kit (Clontech laboratories, Inc.) according to the manufacturer’s instructions. For obtaining full-length cDNA sequence, new gene-specific sense and antisense primers were designed (Table 1). The primers were used to PCR for obtaining full-length cDNA sequence. Nucleotide sequences and deduced amino acid sequence aligned with their respective homologues using Genetyx 7.0 software (GENETYX Corporation, Tokyo, Japan) and sequence alignment editor (BioEdit) (Hall 2011).
The phylogenetic tree was constructed using the Neighbor-Joining Method by MEGA6 (Tamura et al. 2013). Different DNA and protein sequences in Ensembl sequence database were used to carry out phylogenetic tree generation, sequence alignments, and database searching (Additional file 1) (Flicek et al. 2011).
The tissue distribution of PoARF1b in different tissues was measured by RT-qPCR using a LightCycler 480 Real-Time PCR System (Roche, Mannheim, Germany) with LightCycler 480 SYBR green master I (Roche). The total RNA was extracted from the brain, gullet, eye, heart, stomach, liver, kidney, spleen, pyloric ceca, muscle, intestine, and gill from healthy P. olivaceus specimens. cDNA was synthesized with random hexamer primers and oligo(dT)18 using the PrimeScript™ 1st strand cDNA Synthesis Kit (TaKaRa), according to the manufacturer’s instructions. The specific primer for internal control was used 18s rRNA (Table 1) (Ahn et al. 2008). The quantitative real-time PCR followed program: pre-incubation at 95 °C for 5 min, 45 cycles at 95 °C for 10 s, 60 °C for 10 s, and 72 °C for 10 s. The qPCR reaction mixture consists of the following elements: 10 μl of 2× SYBR (Roche), 7.5 μl of SYBR water (Roche), 1 μl each of sense and antisense primers, and 0.5 μl diluted first-strand cDNA (diluted at 1:20). The ΔΔCt method was applied to calculate the data, and 2−ΔΔCt method was applied to calculate the relative quantitative value (Giulietti et al. 2001).
All qPCR data were statistically analyzed using SPSS 21 program (SPSS, Chicago, IL, USA). One-way ANOVA was used to study the PoARF1b expression, followed by Duncan’s Multiple Range test. A p value with p < 0.05 was considered to be significant (Sokal and Rohlf 1969).
Hirame natural embryo cell line (HINAE) was grown in Leibovitz’s L-15 medium (Gibco BRL, Grand Island, NY) containing 10% fetal bovine serum (Gibco) and 1% antibiotics (Gibco) at 20 °C (Kasai and Yoshimizu 2001). The transfection was performed using PolyPlus (JetPrime, New York, NY, USA) kit to transient transfection of PoARF1b and its mutants according to the manufacturer’s instructions in six-well test plates. The PoARF1b and mutants were observed by EGFP fluorescence signal under confocal microscopy after 48 h post-transfection.
PoARF1b(T30N) and PoARF1b(Q70L) were performed using QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies), according to the manufacturer’s instructions (Wang and Malcolm 1999). For PoARF1b mutants, we used specific primers (Table 1). The pEGFP-C1 (Clontech) was used to construct the green fluorescent protein-fused PoARF1b and PoARF1b mutants.
The Golgi body in HINAE was stained using GOLGI ID® Green assay kit containing a Golgi apparatus-selective dye.
Results and Discussion
To identify the initial sequence of PoARF1b, we obtained databases of other ARF1b by using Ensembl sequence data. These sequences were used to design the forward and reverse primers (Table 1). The initial sequence was obtained from PCR amplification of olive flounder cDNA including the brain, eye, gullet, heart, liver, muscle, stomach, kidney, spleen, pyloric ceca, intestine, and gill. The partial sequence was used for isolation of full-length flounder ARF1b by using 3′ and 5′ GeneRace with flounder ARF1b specific primers (Table 1). As the result, the full nucleotide sequence of PoARF1b is 1677 bp (GenBank accession no. KX668134).
The sequence comprised a 108 bp 5′-untranslated region (5′-UTR), a 544 bp coding region, and a 1025 bp 3′-untranslated region (3′-UTR). Also, the PoARF1b has 180 amino acid residues and the molecular weight is approximately 20,561 Da (Fig. 1a). PoARF1b contains GTP-binding motif, switch 1 and 2 regions (Fig. 1a) (Pasqualato et al. 2002). The GTP-binding motif is shaded with gray box and conserved sequence in other ARFs. The switch 1 and 2 regions were indicated with blue and red letters. The switch regions were significantly assumed to conformational change classical structural GDP/GTP switch that bind tightly to GTP but poorly or not at all to the GDP nucleotide (Pasqualato et al. 2002).

The aligned amino acid sequence appeared in PoARF1b had well conserved domains such as GTP-binding motif, switch 1 and 2 regions, and shared the high homology with ARFs from other species (Fig. 1b). It showed 90% homology with ARF1b from Takifugu rubripes.
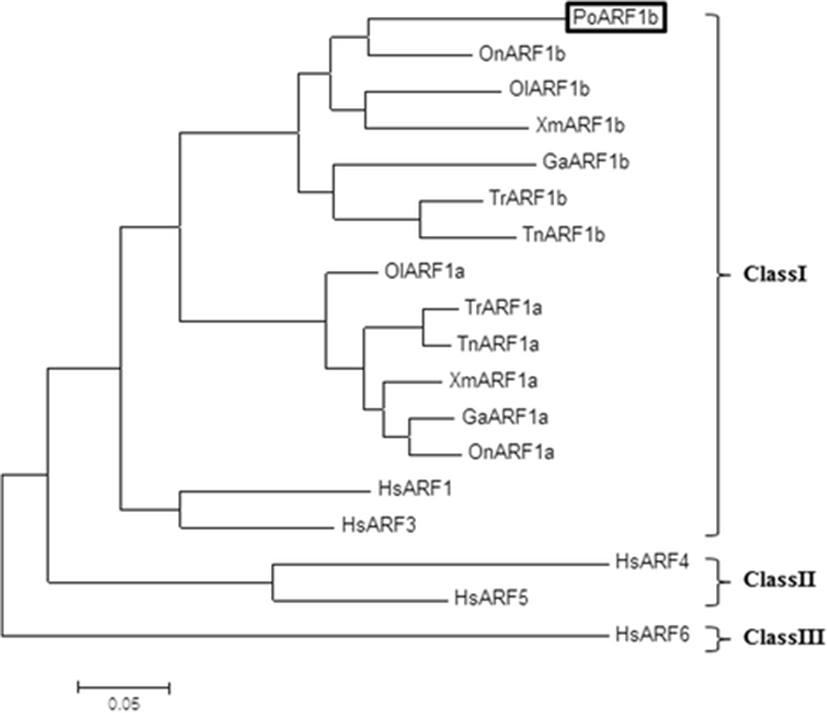
To determine the evolutionary relationship of PoARF1b with other ARFs, phylogenetic tree was performed using the Ensembl sequence data using the Neighbor-Joining Method by MEGA (version 6) with bootstrapping 2000 times (Flicek et al. 2011; Tamura et al. 2013). The result of phylogenetic tree was contained fish which was grouped with the tetrapod and human. The ARF tree consists of three major groups: (i) class I, (ii) class II, and (iii) class III. This result indicated that PoARF1b is closely related to ARF1b of class I (Fig. 2).
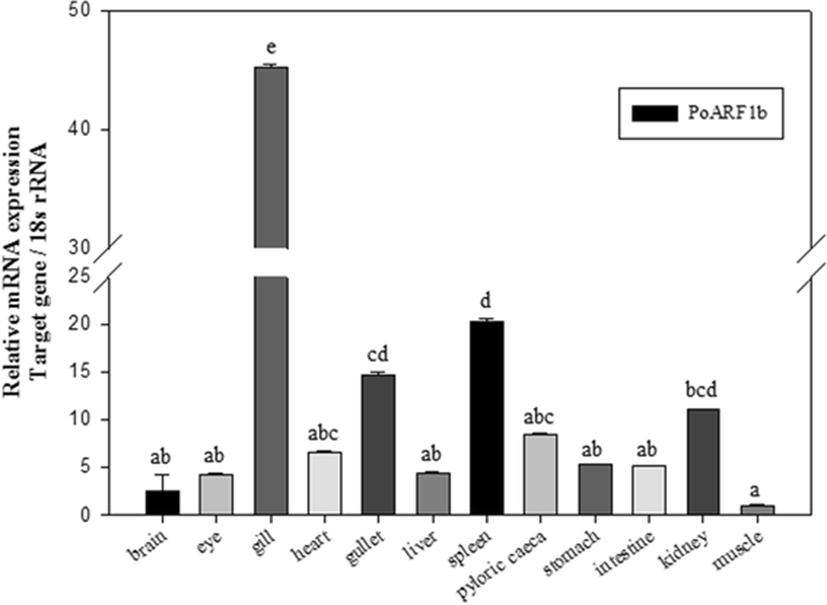
Real-time PCR showed tissue distribution of PoARF1b. The results of qPCR-described PoARF1b was expressed for all mRNA transcripts in different organs which include the brain, gullet, eye, heart, stomach, liver, kidney, spleen, pyloric ceca, muscle, intestine, and gill (Fig. 3). The expression of the PoARF1b gene was the highest level in the gill and the lowest level in the muscle.
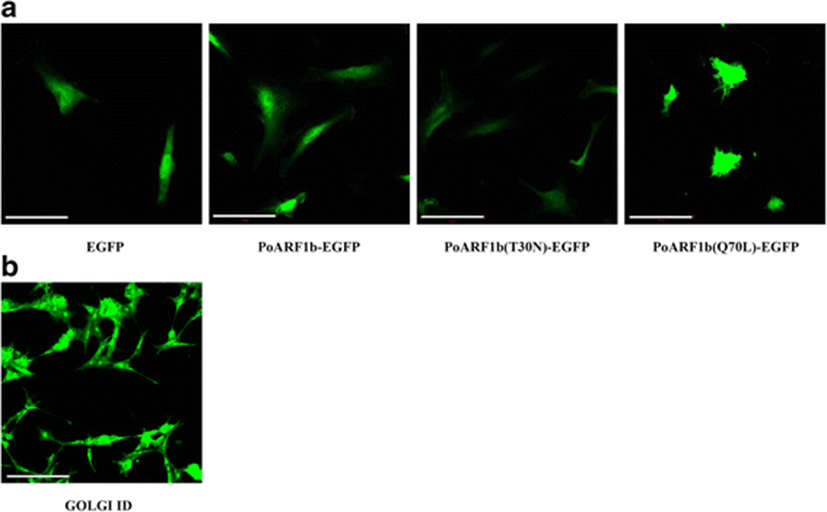
To determine the distribution of PoARF1b, PoARF1b, and PoARF1b mutants, those were constructed into pEGFP-C1 (Clontech) and transfected into HINAE cell. The punctuate morphology of PoARF1b-EGFP resembled the Golgi complex distribution in HINAE cells (Fig. 4b). This result examines PoARF1b that may act in the Golgi body like as human ARF1 complexed with GDP (Amor et al. 1994). Also, PoARF1b mutants examine the distribution which depends on each GDP or GTP-binding form. The mutants designed PoARF1b(T30N) and PoARF1b(Q70L) (Fig. 4a), according to other reports (Chavrier and Goud 1999; Teal et al. 1994). PoARF1b(T30N) was designed by exchanging the Thr amino acid at position 30 with Asn amino acid. It was expected to function in a dominant-negative manner and retain the GDP-binding form. PoARF1b(Q70L) was designed by replacing the Gln at position 70 with Leu. It was expected to function in a dominant-positive manner and keep the GTP-binding form. The result of PoARF1b(T30N) showed a clearly disassembled punctuate morphology (Fig. 4a). When PoARF1b is in a GDP-binding form, it examines to disassociate from the Golgi complex. On the other hand, the result of PoARF1b(Q70L) examined more expanded morphology than that of normal PoARF1b and PoARF1b(T30N) (Fig. 4a). When PoARF1b is in a GTP-binding form, it shows to associate from the Golgi complex.
Conclusion
The small GTPases regulate multiple signaling processes including cell growth, survival, and differentiation (Johnson and Chen 2012). The ARF1 function of the Golgi complex may be important and plays a significant role in the secretory pathway (Radhakrishna and Donaldson 1997). In this paper, Paralichthys olivaceus ARF (PoARF) was cloned. The deduced amino acid sequence of PoARF contains the GTP-binding motif, and the switch 1 and 2 regions are present as mammalian ARF. The PoARF is highly conserved in the other amino acid sequences from teleosts and humans. The PoARF is indicated from approximately 76 to 85% of the overall identities from the other ARF isozymes (data not shown). PoARF shares approximately 85% with the identity of Oreochromis niloticus ARF1b (OnARF1b) and approximately 79% with the identity of Gasterosteus aculeatus ARF1b (GaARF1b). Also, PoARF shares 76% with the identity of Homo sapiens ARF1 (HsARF1). In addition, the phylogenetic tree showed that PoARF is more closely related to ARF1b than ARF1a. These results indicate that PoARF is PoARF1b. OnARF1b, which shares a high percentage with the identity of PoARF1b, shares 76% with the identity of HsARF1.
As is known, the PoARF1b is expressed in all of the tissues of the olive flounder. The PoARF1b mRNA has a high expression level in the gill and a low expression level in the muscle. This finding resembles the ARF1 expression from the shrimp (Marsupenaeus japonicus), which shows the lowest expression level in the muscle (Ma et al. 2010). It will be necessary to further study why such an outcome.
The Golgi-binding distribution of PoARF1b depends on the GTP- or GDP-bound state. The PoARF1b-EGFP showed a punctuate morphology that resembles the morphology of the Golgi body in HINAE cells (Fig. 4). The GOLGI ID of the Golgi bodies can be detected using a microscopy.
PoARF1b(T30N) showed a clearly disassembled punctuate morphology, and PoARF1b(Q70L) showed a more expanded morphology; these results resemble those of the mammalian ARF1. The results of this study indicate that PoARF1b functions within the Golgi complex.
Further studies are needed to be carried out to explain the highest expression of PoARF1b in gill.