Background
For the adaptation to aquatic locomotion, teleosts exhibit the distinct separation of muscle fiber types (slow-twitch fibers to red muscle and fast-twitch fibers to white muscle) into discrete layers in their striated muscles, which has not been seen in other amniote vertebrates (Watabe 1999; Kiessling et al. 2006). White muscle (glycolytic, anaerobic metabolism; for fast-start, burst swimming) and red muscle (oxidative metabolism; for slow, sustained movement) occupy distinct axial regions (Kiessling et al. 2006; Chong et al. 2009). Another striking difference in a skeletal musculature between fish and amniote vertebrates is the indeterminate growth pattern of fish muscles. Most teleosts display a continued growth throughout most of their lifespan, with body size and muscle mass increasing, albeit at a slow rate until mortality or senescence occurs (Johnston et al. 2011). Muscle growth in teleosts is signified by the combined contribution of a long-lasting hyperplasia (increase in fiber number) as well as the hypertrophy (increase in fiber size), which is hardly observable in many other higher vertebrates (Kiessling et al. 2006; Johnston et al. 2011). Furthermore, metabolic and contractile characteristics of teleost muscles also represent significant plasticity and flexibility with regard to not only environmental conditions (e.g., temperature (Hall et al. 2003; Johnston et al. 2008; Stickland et al. 1988; Johnston 2006)) but also reproductive status (e.g., gonad maturation (Johnston et al. 2011; Mathana et al. 2012)).
Taken together, teleosts have developed unique or specialized features in the architecture and physiology of muscles during their evolutionary history, and such adaptive processes undoubtedly have resulted in modifications of the genetic pathways modulating muscle growth and functions in teleosts, as compared to terrestrial vertebrates (Morita 2000; Mudalige et al. 2007). For this reason, identification of genetic determinants for muscle-specific genes from teleosts may be an important basis to study the evolutionary diversification of the musculature in the vertebrate lineage. Also, from an aquaculture viewpoint, myogenic genes are potentially of biotechnological interest from the perspective of growth regulation and quality control of the flesh product. Understanding these key genes could provide the base in order to develop future programs involving genetic manipulation and selection of desired growth trait(s) in commercially important fish species (Johnston et al. 2008).
Actins are one of major components of muscle tissues to compose the myofibrils of musculature. As an evolutionary conserved protein, actins play essential roles in maintaining cytoskeletal structure, cellular mobility, cell division and differentiation, intracellular movement, and contractile processes, which are associated with a wide spectrum of physiological aspects in vertebrates (Perrin and Ervasti 2010). The actin multigene family has been known to be evolved through duplication and divergence from a common ancestral gene, leading diverse isoforms (and subisoforms) in the extant actin genes (Miwa et al. 1991). Although actin isoforms share remarkably high sequence identity one another, each isoform exhibits distinct regulation pattern for its spatial and temporal expression (Perrin and Ervasti 2010). Further, teleosts have been known to retain multiple paralog copies of actin isoform genes depending on taxonomic lineages as a result of a whole-genome duplication (WGD) event (Kim and Nam 2009; Glasauer and Neuhauss 2014). It potentially implies that certain subfunctionalization and/or neofunctionalization might have occurred in teleostean actin isoform groups (Venhatesh et al. 1996). Muscle actins (alpha actins) in teleosts can be broadly classified into three primary types, α-skeletal, α-cardiac, and α-smooth muscle actins. Alpha-skeletal (ACTA1) and α-cardiac (ACTC1) actins are expressed in striated muscles, importantly involved in locomotory action of skeletal muscles and cardiac contractility, respectively. On the other hand, α-smooth muscle actin (ACTA2) is expressed in vascular and visceral smooth muscles (Kim and Nam 2009; Venhatesh et al. 1996). Mostly from mammalian studies, the expression of these three actin isoforms has been known to be regulated through both isoform-specific and cooperative ways depending on development stages and adult tissues (Perrin and Ervasti 2010; Bertola et al. 2008). However, in contrast to rich information on mammalian counterparts, the comparative expression profiles of the muscle actin isoforms in a given single species have remained to be further explored in teleosts.
Mud loach (Misgurnus mizolepis Günther; Cypriniformes) is an aquaculture-relevant food fish in Korea (Kim et al. 1994). This species has various advantageous merits as a candidate model system for muscle research in teleosts. As a model fish, it possesses general merits such as easiness of laboratory maintenance, transparent embryonic development, and relatively short generation time (Nam et al. 2000). Besides, mud loach undergoes characteristic changes in locomotory behavior during early larval development, suggesting that this species is a good subject to visualize the interrelationships between the differentiation of different muscle types and larval swimming behavior. Furthermore, with this species, dramatically induced muscle hyperplasia has been demonstrated by a GH-transgenesis (Nam et al. 2001; Nam et al. 2008), which can provide useful platform to examine differential regulation of genetic pathways associated with muscle growth in hyperplasia/hypertrophy-accelerated fishes. Taken together, our long-term goal is to list this species as a research model for myogenesis of teleosts. This study, as an initial step toward our goal, was aimed to isolate genetic determinants for three representative muscle actins (skeletal, cardiac, and smooth muscle types) from mud loach and to examine the isoform-dependent expression patterns in adult tissues and developing embryos and larvae.
Methods
From next-generation sequencing (NGS) database of mud loach whole fry transcriptome (unpublished data), expressed sequence tag clones showing significant homology to previously known vertebrate muscle actins were collected. The fragment sequences were assembled into contigs using Sequencher software (Gene Codes, Ann Arbor, MI, USA) and subjected to NCBI GenBank BLASTx search to verify their potential annotations as muscle alpha actin members. From this pilot analysis, three distinct actin isoforms were identified to be potentially designated α-skeletal, α-cardiac, and α-smooth muscle actins, respectively.
In order to get full-length complementary DNA (cDNA) for each isoform, vectorette polymerase chain reaction (PCR) was conducted to both 5′- and 3′-directions using the excised phagemid stock of the mud loach fry whole body cDNA library (Agilent, Santa Clara CA, USA) as a template for PCR amplification. Oligonucleotides used in this study were listed in Additional file 1: Table S1. PCR-isolated fragments of each isoform were cloned into pGEM-T easy vector (Promega, Madison, WI, USA), sequenced and assembled into contigs. Based on the contig sequence for each actin isoform, full-length continuous fragment of each isoform was re-isolated from mud loach total RNA (mixed total RNA from muscles and internal organs) by reverse transcription-PCR (RT-PCR). Amplified RT-PCR products were purified and sequenced directly in order to determine the representative cDNA sequence for each α-actin isoform.
Based on the cDNA sequence, genomic gene of each isoform was PCR-amplified from a mud loach genomic DNA template prepared with caudal fin. Amplified fragment(s) for each isoform gene was directly sequenced at both directions by primer walking method. From the genomic gene sequence, the genome walking to 5′-upstream region was carried out in order to identify putative non-translated exon. Genome walking was conducted using Universal Genome Walker Kit (Clontech Laboratories Inc., Mountain View, CA, USA) according to the manufacturer’s instructions. Depending on isoform genes, two to five rounds of genome walking were carried out. Assembled 5′-upstream region for each isoform was re-isolated together with its downstream coding region in a continuous fragment, and finally, the representative sequence of each isoform gene was determined by direct sequencing.
Open reading frame (ORF) in each isoform cDNA was predicted using the ORF finder tool in NCBI page (https://www.ncbi.nlm.nih.gov/orffinder/), and the ORF sequence was deduced into amino acid sequence using the same tool. Homology of each isoform to previously known orthologs was examined by BLAST searches against NCBI GenBank database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Multiple sequence alignments of nucleotide and amino acid sequences were carried out using the ClustalW tool (http://www.genome.jp/tools-bin/clustalw). Physicochemical parameters of protein (with deduced amino acid sequence) were computed using ExPASy ProtParam tool (http://web.expasy.org/protparam/). Phylogenetic analysis of actin isoforms was conducted by MEGA 7 (ver. 7.0.26) software using neighbor-joining algorithm (http://www.megasoftware.net/). In order to compare genomic structure of teleost actins, actin genes were searched against Gene database in NCBI (https://www.ncbi.nlm.nih.gov/gene/), and genomic context of each output was filed for further comparison.
Mud loach specimens used in this study were laboratory-bred strains that have been maintained in Experimental Fish Culture Station, Pukyong National University, Busan, South Korea. In order to examine the expression pattern of actin isoforms in adult tissues, 12 normally grown healthy individuals (six females and six males; average body weight [BW] = 30.1 ± 3.5 g) were selected. From each individual, the brain, eye, fin (caudal), gill, heart, intestine, kidney, liver, muscle (skeletal muscle), spleen, skin, and gonad (ovary or testis) were surgically removed. Upon removal, tissues were immediately frozen on dry ice and stored at − 80 °C deep freezer until used for RNA extraction.
In order to prepare embryo and larvae samples, artificial induced spawning was performed according to the methods described previously (Kim et al. 1994; Nam et al. 2001). Briefly, carp pituitary extract (Sigma-Aldrich, St. Louis, MO, USA) was delivered to six female and six male broodfish via intraperitoneal injection at the dose levels of 10 μg/g BW and 2 μg/g BW, respectively. Pooled eggs (from three females based on egg quality) were divided into three egg batches. Each egg batch was inseminated by sperm (saline-diluted) from each of three individual males in order to prepare three fertilized egg groups. Insemination was done using wet method. Fertilized eggs were rinsed with clean tap water (1-μm-filtered) and placed on an incubator at 25 ± 1 °C with a constant aeration to adjust the dissolved oxygen level to be 7 ± 1 ppm throughout the experiment. Fertilization rates and hatching success of the each replicate group were higher than 98 and 85%, respectively. During the embryonic development, approximately 300 embryos were randomly sampled at 0 (just fertilized), 2, 4, 6, 8, 12, 16, 20, 24, and 28 (hatch-out) hours post fertilization (HPF). Information on the embryogenesis of mud loach can be referred to previous report (Kim et al. 1987). After hatching, the hatched larvae from each fertilization group were transferred to one of three larval incubation tanks (each 60-L rectangular plastic container) equipped with thermostat-assisted heater and aeration apparatus. Larvae were reared at the same temperature as above. After yolk sac absorption (around 60 h after hatch), larvae were fed with 100-μm commercial diet powder for flounder larvae (Woosung Feed Cor., Korea) and live Artemia nauplii (INVE Aquaculture Inc., Salt Lake City, UT, USA). During the larval rearing period, about 100 larvae from each replicate group were randomly sampled every day until 7 days post-hatching (DPH). Sampled larvae were frozen on dry ice and kept in deep freezer until used for RNA extraction.
Total RNA was extracted using TriPure Reagent (Roche Applied Science, Mannheim, Germany) and RNeasy Plus Mini Kit (Qiagen, Hilden, Germany) according to manufacturers’ recommendations. Quality and quantity of total RNA extracted from tissues were verified by MOPS-formaldehyde agarose gel electrophoresis and NanoDrop Microvolume spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Two microgram of total RNA from each sample was reverse transcribed into cDNA by using Omniscript® Reverse Transcription Kit (Qiagen, Germany) according to the manufacturer’s protocol. During reverse transcription (RT) reaction, a mud loach 18S rRNA reverse primer was included in order to prepare the normalization control for quantitative reverse transcription PCR (RT-qPCR) assay (Nam et al. 2011). The synthesized cDNAs were fourfold diluted with sterile distilled water, and 2 μL of diluted cDNA was used for each quantitative PCR (qPCR) amplification. PCR was run with LightCyler480® II Real-Time PCR System (Roche Applied Science, Germany) and LightCycler® DNA Master SYBR Green I (Roche Applied Science, Germany). Primer pairs for each actin isoform and 18S rRNA normalization control were confirmed to amplify the specific band based on the melting curve analysis after the thermal cycling. PCR efficiency of each primer pair was validated to be higher than at least 92% based on the standard curve for each gene. Quantitative PCR assay of each cDNA sample was carried out in triplicates. After each thermal cycle, amplification signals were analyzed with LightCyler480® II Software (ver. 1.5; Roche Applied Science, Germany). Relative mRNA expression levels of each actin isoform in each tissue and developmental stages were estimated by ΔCt method based on the normalization against its own level of 18S rRNA control (Schmittgen and Livak 2008). Significantly different expression levels among tissues and developmental stages were addressed by one-way ANOVA followed by Duncan’s multiple ranged test at the level of P = 0.05.
Results and discussion
The three α-actin isoforms isolated from this species were designated, ACTA1 [1268 bp excluding the poly(A+) tail], ACTC1 (1273 bp), and ACTA2 (1607 bp) based on the homology search and molecular phylogeny (see below). The three isoform cDNAs showed the same stop codon (TAA). Canonical putative polyadenylation signal (AATAAA) was predicted in the three isoform, 23–26 bp prior to the poly(A+) tail. However, ACTA2 showed an additional polyadenylation signal (same AATAAA sequence) in the vicinity of the poly(A+) tail (10 bp prior to the tail). The 5′- and 3′-untranslated regions (UTRs) of ACTA2 are relatively longer than that of ACTA1, similarly reported in other cypriniform fish (Kim and Nam 2009). All of the three isoforms revealed a single ORF (each 1131 bp excluding stop codon) to encode the same number of amino acids (377 amino acids, including the first two Met-Cys residues known to be processed after translation). Sequences of the three isoforms are available in GenBank under the accession numbers KX347544 (ACTA1), KX347545 (ACTC1), and KX347546 (ACTA2). Summarized information on the sequence characteristics of mud loach α-actin isoforms is also provided in Additional file 2: Table S2.
Three mud loach α-actin isoforms represented highly similar calculated molecular masses (41,959–41,995 Da) and theoretical isoelectric points (pI = 5.22–5.23). Sequence identity among them at the amino acid level was at least more than 98%, in which there were only eight non-conservative amino acid substitutions out of 377 residues (Fig. 1). Thereby, our sequence data consistently confirm the known appraisal on the divergence of actin isoform genes from a common ancestral form through duplication events (Miwa et al. 1991; Venhatesh et al. 1996). In addition, from the multiple sequence alignments of each isoform with its representative orthologs also showed a considerably high degree of amino acid sequence identity ranging from 97 to 100% among species (data not shown).
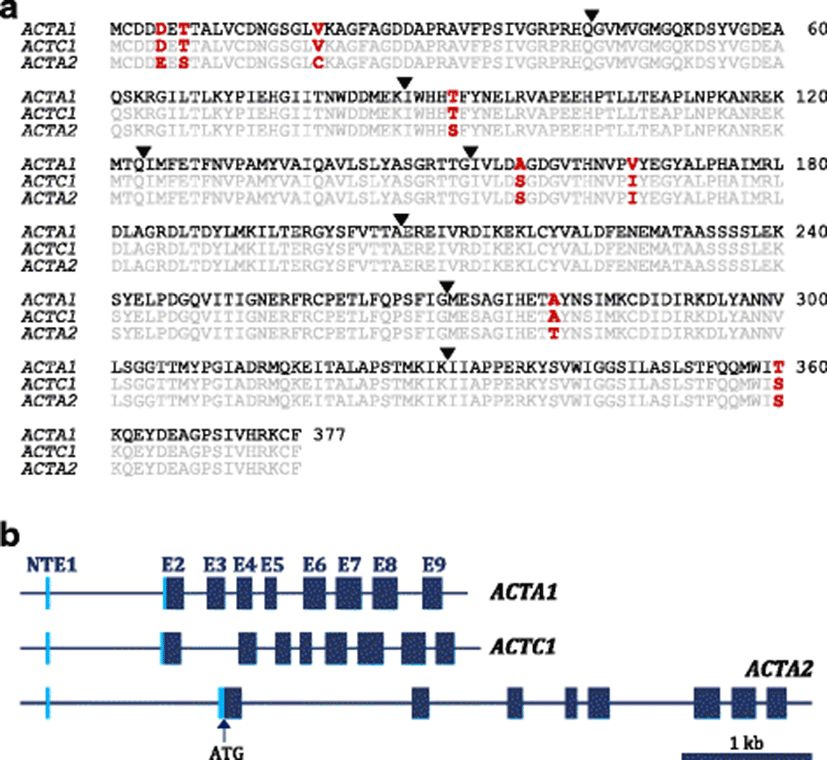
A closer look into the sequence alignment among the three mud loach α-actin isoforms indicates that substitution patterns in several non-conserved residues are similar with those observed in other teleost actin isoforms. For example, the alanine/serine substitution found at position 157 (Ala/Ser-157) is also found (1) between pufferfish α-skeletal actin isoform 1 and isoform 2 (Venhatesh et al. 1996), (2) between rattail fish (Coryphaenoides acrolepis and C. cinereus) α-skeletal actin isoforms (Morita 2000), and also (3) between the two types of Leporinus macrocephalus skeletal α-actin isoforms (Alves-Costa et al. 2015). In particular, the substitution in L. macrocephalus α-actin isoforms was observed between actin isoforms isolated respectively from white muscle (Ser-157) and red (Ala-157) muscle (Alves-Costa et al. 2015). This amino acid substitution occurs in the vicinity of actin ATP-binding site and has been proposed to influence the actin’s affinity to ATP (also to divalent cation) based on the assumption that Ser has a higher hydrogen bonding capacity than Ala (Morita 2000; Alves-Costa et al. 2015). Affinity of actin to ATP has also been reported to affect the thermal stability of fish actins (more stable in ATP-bound forms) extracted from myofibril powder (Torigai and Konno 1997). Similarly, the substitutions among mud loach α-actin isoforms Thr/Ser-91, Val/Ile-167, and Ala/Thr-280 are observed among subisoforms of pufferfish α-skeletal actin (Venhatesh et al. 1996).
In order to designate the nomenclature of three α-actin isoforms cloned in mud loach, a molecular phylogenetic analysis was carried out with the taxa sampled from representative teleost and amniote species (Fig. 2). Molecular phylogenetic analysis resulted in a monophyletic status of three α-actin isoforms (skeletal, cardiac, and smooth muscle forms), although the topology was not supported by the high bootstrap value. Teleost α-skeletal actins and α-smooth muscle actins formed the two monophyletic clades, where mud loach ACTA1 and ACTA2 branched off, respectively, in each of the two clusters (skeletal and cardiac). Mud loach ACTA1 isoform showed the closest phylogenetic affiliation to Cyprinus carpio ACTA1, while mud loach ACTA2 displayed the closest relationship with Hemibarbus mylodon and Danio rerio ACTA2 orthologs. However, in the present analysis, teleost α-cardiac actins did not form a monophyletic clade. Rather, they were divided into two subclades, in which the C. carpio and D. rerio ACTC1s formed a small, their own subclade, although the node did not receive the statistical support. Furthermore, the mud loach ACTC1 was not closely affiliated with orthologs from the two cypriniform species (C. carpio and D. rerio); instead, it is closely related with α-cardiac orthologs from distantly related species such as Takifugu rubripes and Oreochromis niloticus. Because we did not sample all the presumed α-cardiac actins from the teleosts according to our primary objective of molecular phylogeny (i.e., type identification of three mud loach isoforms for their proper nomenclatures), extensive reconstructions of the tree with much more complete sequence sampling might also be needed in future. Also, the further mining of additional paralog actin isoforms from the mud loach genome might be helpful to explain the reason behind this unexpected finding.
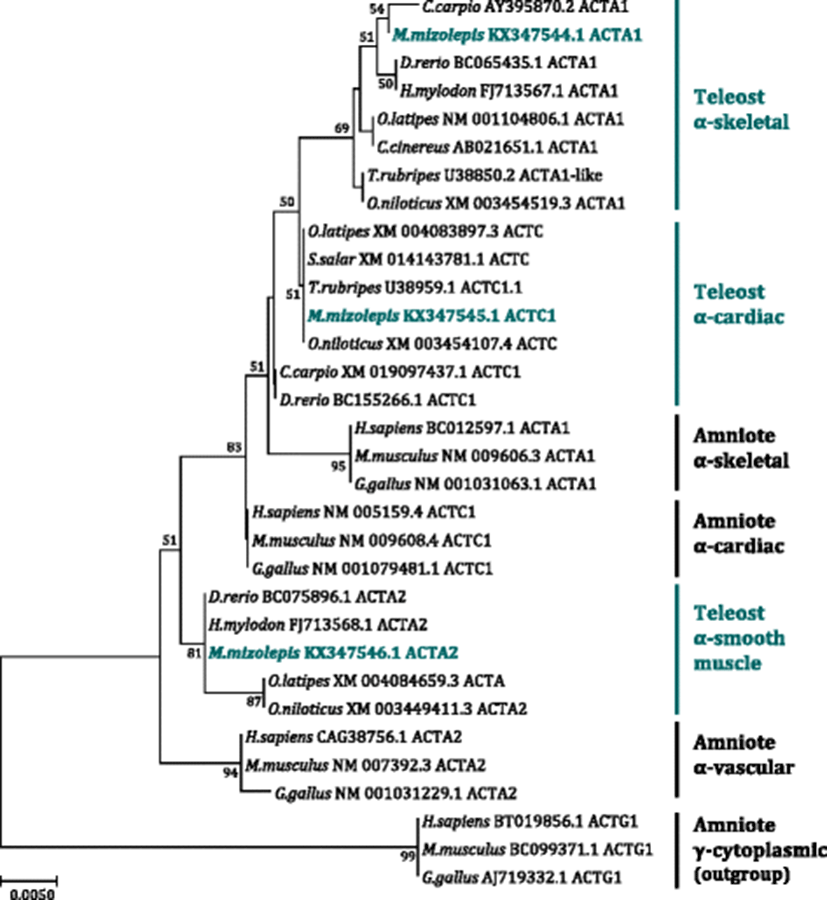
Inconsistency between phylogenetic affiliation and previously known taxonomic appraisal of the species has been reported in previous attempt on the reconstruction of phylogenetic tree of teleost muscle actins, in which complex pattern of phylogenetic relationships would be apparent due to significant gene duplications and diversifications of actin paralogs in teleost genomes (Kim and Nam 2009). Gene duplications following differential subfunctionalization(s) seemed to have occurred in an isoform- and species (or lineage)-dependent fashion, as particularly noticed in Salmoniformes, Gadiformes, Cypriniformes, and Tetraodontiformes (Johnston et al. 2008; Kim and Nam 2009). Furthermore, certain muscle actin isoforms observed in these taxa exhibited no clear affiliation to currently established orthology in vertebrate actin multigene family, making the phylogenetic explanation more complicated (Kim and Nam 2009; Venhatesh et al. 1996). Whatever the evolutionary mechanism(s), teleostean muscle actins have experienced different evolutionary history in comparison to amniote counterparts, and diversified paralogs could be the potential source of muscle plasticity to interact with their environmental changes or variations.
From the genomic cloning, the coding sequence of each isoform gene was clearly matched to its corresponding cDNA counterpart, and the GT/AG exon-intron splicing rule was well conserved at each boundary region. Genomic architectures (exon-intron organization) of the three actin isoforms were found to be identical (eight translated exons), again strongly suggesting that they have derived from a common ancestral gene that had gone through duplications and conversions. Based on the alignment of cDNA and genomic sequences, each of three mud loach α-actin isoforms was proven to possess a non-translated exon 1 (NTE1). Lengths of NTE1s found in ACTA1 (KX347541), ACTC1 (KX347542), and ACTA2 (KX347543) were 24, 22, and 33 bp, respectively. Although the presence of NTE1 has not been extensively characterized in fish α-actin genes, our bioinformatic survey has shown that similar NTE1s exist in many other teleost α-actin genes (data not shown) as reported in human actin genes (Miwa et al. 1991). Besides the NTE1, three α-actin isoform genes from mud loach represented equally eight translated exons (exon E2 to exon E9) and the length of coding sequence in each translated exon (E2–E9) was perfectly identical among the three isoform genes (129, 129, 111, 85, 162, 192, 182, and 144 bp, respectively for E2 to E9). On the other hand, lengths of non-coding, intronic lengths are variable among the three isoform genes (Fig. 1; Additional file 2: Table S2).
Actins are one of the most conserved protein groups, and extremely high sequence similarly of actin proteins are commonly found among almost eukaryotic organisms. However, in contrast to such a homogenous feature at protein level, genomic exon-intron organization of actin genes (i.e., number of exons) has been known to be variable among species and/or taxonomic lineages (Bertola et al. 2008; Kusakabe et al. 1999). For example, most mammalian α-skeletal and α-cardiac actin genes reveal six translated exons with the conserved intron positions, while α-vascular and γ-enteric smooth muscle actin genes exhibit eight translated exons in mammals. Ascidian muscle actins show four or five exons depending on larval and adult muscle types. Many insect species lost all introns in their α-skeletal actin genes. Caenorhabditis elegans (nematode) have three or four exons in its actin genes (Bertola et al. 2008; Kusakabe et al. 1999). Further, teleost species, which have experienced WGD in their evolutionary history, are known to retain more diverse paralogs in actin multigene family. With this context, the gene architecture (exon-intron organization) of teleost actins is thought to be relatively more diversified, compared to ones established in other vertebrates including mammals. However, barring only a few examples, the comparative information on genomic organization patterns of teleost actin genes has not been sufficiently described.
As mentioned above, mud loach displays a uniform pattern of genomic organization (eight translated exons interrupted by seven introns) for all the three actin paralogs. However, our bioinformatic survey to other teleost actin loci strongly indicates that such a uniform pattern observed in mud loach may not be conserved in all other teleostean genomes. Rather, the exon-intron organization is found to be highly species- and isoform-dependent in the teleost lineage (Additional file 3: Table S3). For ACTA1 (skeletal actin), the gene organization with six translated exons is the most frequently found form, which is corresponding to the organization pattern of all known mammalian α-skeletal actin genes. ACTA1s with six translated exons are exemplified in channel catfish Ictalurus punctatus (GenBank Gene ID = 108263396), pufferfish Takifugu rubripes (Gene ID = 101069447), large yellow croaker Larimichthys crocea (Gene ID = 104920157), zebrafish Danio rerio (Gene ID = 550445), and tilapia Oreochromis niloticus (Gene ID = 100534413). On the other hand, Japanese flounder Paralichthys olivaceus and common carp Cyprinus carpio represent different numbers (six to eight) of translated exons depending upon paralog copies of ACTA1. Meanwhile, many fish species examined in the present study reveal various paralogs (subisoforms) with differential exon numbers (six to eight) within the ACTC1 isoform (cardiac type). Exceptional species is zebrafish, which highlights only the eight-exon-structured ACTC1 currently. Unlike striated muscle actins (ACTA1 and ACTC1), all the ACTA2 (smooth muscle type) isoforms are comprised of eight translated exons, suggesting teleosts have retained the conserved gene organization pattern for ACTA2 with mammalian ACTA2s (vascular, aorta actins). Taken together, the number of translated exons of actin genes in teleosts is species- and isoform-dependent, in which the organization pattern could also be further diversified among paralog copies within a given isoform. Collectively, the present analysis is strongly suggestive of the complex evolutionary history of actin multigene family in the teleost lineage.
Based on the RT-qPCR assay, mRNA expression patterns of three actin isoforms were examined in adult mud loach tissues (Fig. 3). Mud loach ACTA1 transcripts were detectable in all the tissues examined. As expected, skeletal muscle exhibited the most robust expression of ACTA1 among tissues (P < 0.05). Besides the muscular expression, the intestine, skin, gill, and eye revealed the moderate expression of ACTA1, whereas other remaining tissues displayed weak expression levels (P < 0.05). The lowest expression of ACTA1 was found in the ovary (P < 0.05). On the other hand, ACTC1 transcripts were not clearly detectable in several tissues including the brain, fin, kidney, liver, and gonads under the present RT-qPCR conditions. The heart expressed the highest amount of ACTC1 transcripts. Although skeletal muscle showed the second highest expression level of ACTC1, the expressed amount in skeletal muscle was significantly lower than that in the heart (P < 0.05). Gill and spleen exhibited quite a low expression of ACTC1 in this species, while the eye, intestine, and skin showed a modest expression of ACTC1 (P < 0.05). Meanwhile, the expression pattern of ACTA2 was also ubiquitously observable in all the tissues. The highest expression was found in the intestine, followed by the spleen (P < 0.05). Interestingly, testis revealed an active transcription of ACTA2, of which expression level corresponded to the third highest among all the tissues examined. The heart also expressed a moderate level of ACTA2 transcripts while the other remaining tissues were found to show a weak or minute expression of ACTA2 (P < 0.05).
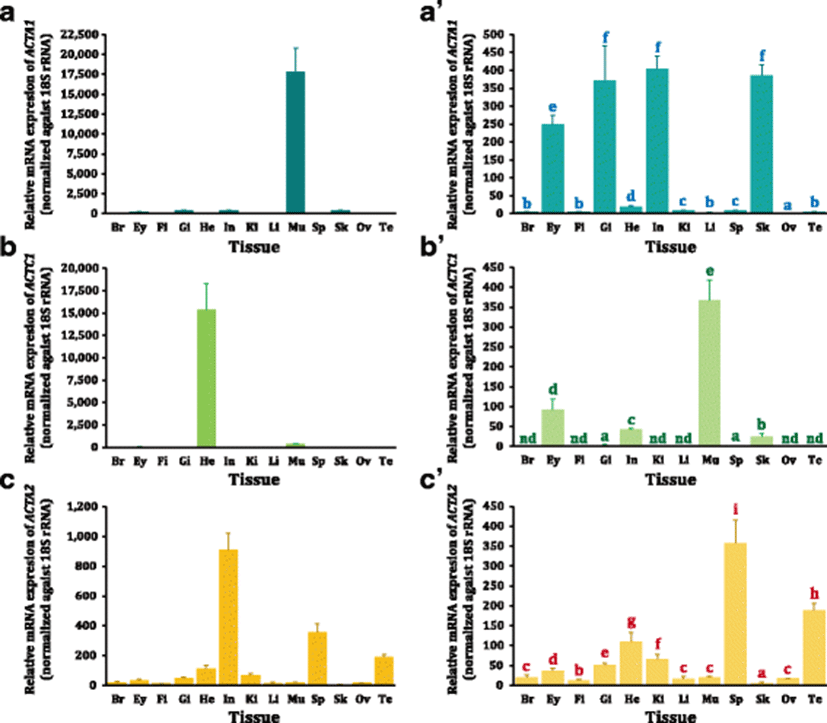
Overall, tissue distribution patterns of the three actin isoform mRNAs observed in the present study are well in agreement with their annotated nomenclatures addressed by phylogenetic analysis, as the highest or predominant expression levels of skeletal (ACTA1)-, cardiac (ACTC1)-, and smooth muscle (ACTA2)-actin genes in the myotomal muscles, heart, and intestine, respectively. Such an isoform-dependent expression pattern indicates a certain subfunctionalization of these three actin isoforms with regard to their tissue-specific roles in the musculature of adult mud loach (Perrin and Ervasti 2010; Bertola et al. 2008). Within the isoform ACTA1, one notable finding is its active transcription in the intestine, which is not generally congruent with the expected pattern of the skeletal muscle actins, since the intestine is a known smooth muscle tissue. Previously, a histological analysis has reported that loach would possess the proximal intestine consisting of intermingled striated and smooth muscle cells (Hara et al. 1989). One plausible, but untested, assumption on this finding (i.e., the considerably active expression of ACTA1 in the intestine) is that this species uses its intestine as an accessory air-breathing organ (Luo et al. 2016), and thereby mud loach may require a unique contractility in order to eject air bubbles from its intestine.
As a main isoform playing essential roles in cardiac contractility, the predominant expression of ACTC1 in the mud loach heart is not surprising. Similar or redundant expression pattern of ACTC1 with ACTA1 (i.e., active expression in skeletal muscle as well as the modest expression in the eye, intestine, and skin) has also been examined previously (Bertola et al. 2008). On the other hand, the vigorous expression of ACTA2 in the intestine is in agreement with its functional involvements mainly in smooth muscle-contained tissues. In mammals, two differentiated isoforms of smooth muscle actins, ACTA2 (vascular/aorta type) and ACTG2 (gastrointestinal type), have been reported (Miwa et al. 1991). However, our peer-review has been suggestive of no clearly characterized ACTG2 in teleosts. Rather, teleosts potentially represented novel and/or anomalous subisoforms/isoforms with little orthology to mammalian actins (Kim and Nam 2009; Venhatesh et al. 1996). A noticeable finding in the expression pattern of mud loach ACTA2 is the prominently high expression in the testis, which is not seen with other two isoforms. The present finding suggests certain putative roles of ACTA2 in the development of testis, which is also similar with previous identification of a testis-specific anomalous actin isoform in the pufferfish (Venhatesh et al. 1996).
Three actin isoforms are differentially modulated during the period of embryonic development and larval early ontogeny in mud loach (Fig. 4). During embryonic development, the onset expression of ACTA1 mRNAs was faintly detected by RT-qPCR at late gastrula stage (8 HPF; characterized by 70% epiboly cover). The expression was gradually increased with the progress of somitogenesis until 24 HPF (corresponding to pre-hatching stage exhibiting more than 30 somites), suggesting the essential roles of ACTA1 in embryonic myogenesis in mud loach embryos (Hall et al. 2003; Xu et al. 2000; Moutou et al. 2001). Afterward, the transcript level of ACTA1 in mud loach embryos was sharply increased until hatch-out and further rigorously until 2 DPH. Then, the expression of ACTA1 was rapidly dropped to the level observed at just hatching (28 HPF) and kept to be relatively constant in subsequent ages. Myogenesis has been known to initiate at an earlier stage of development in teleost embryos than in amniotes; the myogenic precursor cells for slow and fast muscle in fish embryos have been reported to be spatially segregated before somite formation (i.e., as early as gastrulation) (Hirsinger et al. 2004). Hence, the onset expression of ACTA1 transcripts at late gastrula stage in this study is broadly in agreement with the previous observation. The earlier initiation of myogenesis in fish has been proposed to reflect the requirement to generate swimming propulsion at earlier life stages imposed by external fertilization (Johnston et al. 2011). Sharp increase of the expression from 24 DPH might be in relation with embryos’ preparation to develop skeletal muscles for hatching event. After hatch, the mud loach yolk sac larvae rarely swim with the settlement at the bottom. However, in this period also, they seem to continue active transcriptions of skeletal muscle genes including ACTA1 in order to make them fully ready for the start-up of active swimming immediately after yolk sac absorption (i.e., the first time full-scale swimming in their lives) (Fujimoto et al. 2006; Gap et al. 2014). Once larvae are fulfilled with transcripts, ACTA1 transcription is downregulated in a certain degree and then stabilized in subsequent phases. Transition to exogenous feeding after yolk sac absorption usually occur at the period from 2 DPH to 3 DPH at 25 °C in mud loach, which is in accordance with our explanation on the larval ACTA1 gene expression pattern.
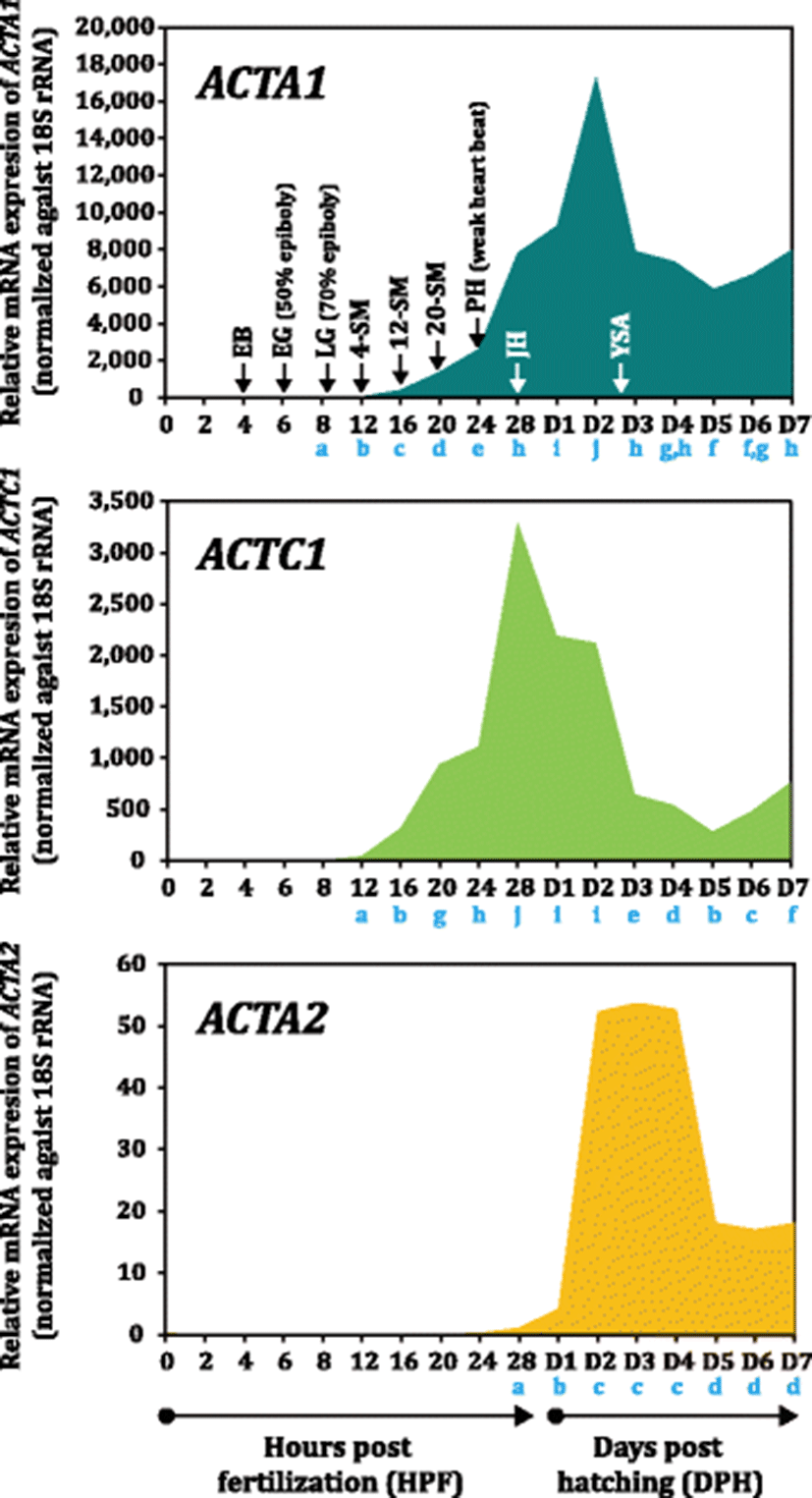
On the other hand, the ACTC1 expression began to be apparent at 12 HPF (4-somite stage), continuously stimulated with the progressive development of embryonic myotomes until 24 HPF and quickly upregulated to reach a sharp peak at 28 HPF (just hatching). Redundant expression of ACTA1 and ACTC1 in the development of striated muscle during embryogenesis is a known phenomenon in vertebrates (Bertola et al. 2008). Although ACTC1 eventually becomes the main actin isoform in the heart in adulthood, it has also been reported to be largely associated with early muscle development (Bertola et al. 2008). A steep increase of ACTC1 expression from the 24 DPH in mud loach embryo is congruent with the initial heart beating and circulation. Afterward, the transcription of ACTC1 was rapidly downregulated until 5 DPH, although there was a lag period of downregulation from 1 DPH to 2 DPH. This is also similar with previous findings, in which the expression of α-cardiac actin would be downregulated and α-skeletal actin becomes the dominant isoform in later developmental stages, eventually accounting for almost proportion of the total striated muscle actins in grown-up vertebrate animals (Sassoon et al. 1988).
Unlike the above two actin isoforms, mRNA expression of ACTA2 was not clearly projected during embryonic development stages, and the initiation of its expression was observed at hatching (at 28 HPF) in a very low amount, which differs the previous demonstration that α-smooth muscle actin would be also expressed during early cardiac and skeletal muscle development in various myofibroblast-like cells in mammalian embryogenesis (Bertola et al. 2008). Currently, it is not clear whether other paralog subisoform(s) of mud loach ACTA2 may participate in the development of early embryonic muscles or not. However, our finding on the absence of ACTA2 transcription in embryogenesis is, at least in part, accordant with the previous study on zebrafish ACTA2 to show that mural cells (main expression site of zebrafish ACTA2) would develop relatively late as compared to ones in mammals (Whitesell et al. 2014). The authors have hypothesized that the slow development of vascular mural cells might be a reflection of the small body size of the zebrafish, and thus, there would be little need to develop contractile smooth muscle at an early embryo stage (Whitesell et al. 2014). In this study, the low level of ACTA2 transcripts in just hatched larvae was slightly increased at 1 DPH; however, soon after, the expression of ACTA2 transcripts was dramatically increased at 2 DPH and kept to be constant until 4 DPH (i.e., immediately following the transition to exogenous feeding). This modulation pattern could be explained by the need for the preparation of smooth muscles with the developments and differentiations of vascular and gastrointestinal organs. Although we did not examine histological ontogenesis of internal organs, initial developments of various smooth muscle-related organs including intestine might occur in that period, as scrutinized in a closely related Misgurnus species (M. anguillicaudatus) (Han et al. 2013; Zhang et al. 2016). Subsequently, ACTA2 transcripts were detected in a decreased level (i.e., at 5 DPH and afterwards). Generally, this pattern might be recognized by a stabilization process after the explosive transcriptions in line with the ontogenic developments of various smooth muscle-relevant organs. However, it may not also be excluded the possibility that ACTA2 transcripts might be diluted in the total RNA templates by various other gene transcripts and dominated ACTA1 transcripts with the increase of body mass in fry. Collectively, the three α-actin isoforms are dynamically modulated during embryo development and larval ontogeny largely in an isoform-dependent fashion.
Conclusion
Three muscle α-actin isoform (ACTA1 for skeletal, ACTC1 for cardiac, and ACTA2 for smooth muscle types) cDNAs and genomic genes were isolated and characterized in mud loach (Misgurnus mizolepis). These three actin isoforms share a high degree of structural homology one another at both protein and genomic levels. However, despite such a high structural homology, these three actin isoforms display an apparent isoform-dependency in tissue and developmental expression patterns, suggesting the functional differentiation of these actin isoforms in mud loach musculature. Overall, the expression pattern of each actin isoform is in accordance with its proposed roles in myogenesis and muscle growth. Data from this study could be a useful basis to designate various future researches regarding myogenic regulation and other muscle-related physiology of this fish species.