Background
Metallothionein (MT) is a metal-binding protein identified in almost living organisms. This housekeeping protein is a multifunctional, non-enzymatic effector playing important roles in various criteria of physiology of organisms under basal and stressed conditions (Carpenè et al. 2007; Mao et al. 2012). Owing to the high inducibility of its expression upon metal exposure, MT has been long given attention as one of core suits for biomarkers to address risks associated with metal-related environmental problems (Sarkar et al. 2006). Besides its fundamental roles in the homeostatic regulation of essential metals and detoxification of trace metals, MT plays important roles in host protective pathways under environmentally or physiologically perturbed conditions (Inoue et al. 2009; Chiaverini and Ley 2010; Lynes et al. 2014).
Benthic mollusks have been proposed as the useful sentinel platforms for biomarker approaches to aquatic and marine environments, in relation with their high bioaccumulation capacity of chemical elements from both water and sediment (Amiard et al. 2006; Geffard et al. 2007; Le et al. 2016). Further, their sedentary nature also makes it possible that biomagnification effects of the pollution could be effectively visualized without a significant consideration of complex migratory factors in the interpretation of bioaccumulation data (Gupta and Singh 2011). Accordingly, the exploitation of genetic determinants of molluskan MTs has been a progressively growing domain in the field of MT researches. To date, a number of previous literatures have claimed that molluskan species should represent a great structural diversity of MT proteins (Jenny et al. 2004; Jenny et al. 2006; Leignel and Laulier 2006). Moreover, some mollusk species particularly including American oyster Crassostrea virginica have shown extraordinarily large-sized MT isoforms comprising of over 100 amino acid (aa) residues (Jenny et al. 2004; Jenny et al. 2006; Tanguy and Moraga 2001), which have not been usually observable in vertebrate orthologs (Blindauer and Leszczyszyn 2010; Serén et al. 2014). Based on this, it has been widely proposed that the mollusks might have undergone evolutionary unique history in the divergence of MT proteins (Serén et al. 2014; Isani and Carpenè 2014; Wang et al. 2014; Jenny et al. 2016). Undoubtedly, structural diversifications would confer functional variations on these molluskan MTs, leading species-specific adaptation to environmental changes. Even yet fully elucidated, the prevalence of diversified isoforms are also likely in relation with significant variabilities and inconsistencies in the responses of molluskan MTs to metal or other stress exposures (in both laboratories and fields) (Amiard et al. 2006; Le et al. 2016).
Evolution of molluskan MTs has been proposed to be fundamentally based on the duplication events of their metal-binding domains. Domain duplication(s) from a common ancestral MT (a singular domain MT) might have given rise to multi-domain structured MTs (Jenny et al. 2004; Palacios et al. 2011). The prototypic MT has been supposed to go through further diversification into various distinguished isoforms in different taxa. This evolutionary theory has been comprehensively addressed with several well-known molluskan models such as oysters, mussels and air-breathing snails (Leignel and Laulier 2006; Jenny et al. 2016; Palacios et al. 2011; Aceto et al. 2011; Leung et al. 2014). However, in contrast to rich information on these popularly studied species, divergent processes of MTs in other mollusk species (in both gastropods and bivalves) have been quite limitedly explored yet. As the research on molluskan MTs progresses, it is becoming increasingly evident that structural variations and deviations of MT domains in this phylum may be significantly larger than previously understood. Recently, there have been considerable efforts on the isolation of novel MTs or MT-like proteins from different molluskan taxa. Now it is not difficult to find certain molluskan MT isoforms of which domain structures are hardly assigned into one of traditionally described categories. Hence, with this viewpoint, we aimed to review the structural diversity of molluskan MT domains with an angle to shed additional light onto the evolutionary path of MT domains in the molluskan lineage.
Status of MT isoform sequence data in public database
Currently the number of publically released MT gene sequences (both genomic and mRNA sequences; https://www.ncbi.nlm.nih.gov/genbank/) from the mollusk species has exceeded over 100 excluding the partial sequences or untrimmed ESTs. Almost all molluskan MT genes have been obtained from bivalves and gastropods, whereas no characterized MT sequence has been reported yet from classes such as Aplacophora, Monoplacophora, Polyplacophora and Scaphopoda. No cloned MT sequence is available in the class Cephalopoda, barring couples of uncharacterized sequences predicted from the octopus (Octopus bimaculoides) genome scaffold. Hence, it is evident that genetic determinants of MT have been yet narrowly explored in the phylum Mollusca.
Many gastropod and bivalve species possess multiple paralogue MT isoforms. American oyster Crassostrea virginica (Ostreidae; Bivalvia) is the top species showing the greatest number of coexisting MT sequences in a single species (n = 17; non-redundant paralogue sequences at aa levels) (Jenny et al. 2004; Jenny et al. 2016); this isoform number is also the highest among all metazoans. Currently, in the Bivalvia class, 34 species belonging to four subclasses (Pteriomorphia, Anomalodesmata, Heteroconchia, and Palaeoheterodonta) have been recorded to reveal MT isoform(s) under GenBank accession codes. However, the species distribution for GenBank-deposited MT sequences in bivalves is highly skewed, in which only a few popularly studied species have dominated MT sequences. More than 50% of the total MT sequences have been from only the top five species belonging to either Mytilidae (mussels) or Ostreidae (oysters). Meanwhile, in the Gastropoda class (the largest and primitive class in the phylum Mollusca), only less than 20 non-redundant full-length MT sequences have been exploited from twelve different species belonging to one of seven families. These gastropod species include air-breathing pulmonate snails belonging to the Heterobranchia (species from four families, Planorbidae, Physidae, Helicidae, and Bradybaenidae), abalones (Haliotidae; Vetigastropoda), a limpet (Fissurellidae; Vetigastropoda), and a periwinkle (Littorinidae; Caenogastropoda). The species information and the accession code for each MT sequence used in this review are referred to in Additional file 1: Table S1.
A number of previous publications have reportedly indicated that most vertebrate MTs including mammalian and teleostean MTs may exhibit well-conserved primary structures. They are typically characterized by common features in polypeptide length (60~65 aa), calculated molecular weight (6~7 kDa), proportion of cysteine (Cys) residues (30~35%; arranged typically as Cys-X-Cys, Cys-X-X-Cys and Cys-Cys where the X is any non-Cys aa), and theoretical pI values (8.0~8.5) (Wang et al. 2014; Capasso et al. 2003; Cho et al. 2005; Cho et al. 2008; Cho et al. 2009) (Additional file 1: Figure S1A). Such a structural homology among vertebrate MTs may also attribute to a considerable degree of functional orthology between vertebrate species. However, it may not apply to the mollusk species. Remarkably variable or deviated scores in the primary structure have been frequently observable in molluskan MTs (Jenny et al. 2004; Tanguy and Moraga 2001; Baršytė et al. 1999; Tanguy et al. 2001) (Additional file 1: Figure S1B). First, molluskan MTs reveal often significant variations in polypeptide lengths among paralogue and orthologue isoforms. Median and mean values for the number of aa residues estimated from all the mollusk MT polypeptide sequences addressed in this study (n = 109) are 73.0 (7.2 kDa) and 80.2 (7.8 kDa) aa, respectively. Overall, these values are higher than that estimated with representative vertebrate MT orthologs. More noticeably in molluskan group, considerable numbers of MTs represent extraordinarily short or long lengths apparently deviated from the mean (and median) value above. Within the Bivalvia class, the length of MT polypeptides ranges from 43 aa (4.2 kDa) to 204 aa (20.3 kDa); noticeably both are recorded as paralogs from a single species C. virginica (Ostreidae) (GenBank accession numbers, AY331700/AY331701 and AY331706, respectively). Other oyster species also display the significant variations in the sizes of MT isoforms. However, the mussel species belonging to the order Mytiloida represents relatively a uniform feature in the range of the MT protein lengths (66~75 aa). Besides the two main bivalve taxa, it is worthy to note unusually long MT isoforms reported from Argopecten irradians (Pectinidae; EF093795, and EU734181) and Pisidium coreanum (Sphaeriidae; GQ268325).
On the other hand, mean and median values for MT polypeptide lengths available for gastropod MTs were 67.5 and 65.0 aa, respectively. Currently, the shortest gastropod MT sequence is found to be Physa acuta (Physidae; GU259686) MT consisting of 59 aa, whereas the longest one is the MT isoform with 100 aa reported in a periwinkle species Littorina littorea (Littorinidae; AY034179). Except these shortest and longest sequences, the polypeptide lengths of other gastropod MTs are found to fall in the range from 64 to 70 aa. Similarly with bivalves, multiple isoform MTs (n = 2~4) have been reported in pulmonate snails belonging to Helicidae or Planorbidae, while marine gastropods including abalones have shown only a single MT isoform; yet still unclear if they have additional MT isoform gene(s) or not (Additional file 1: Table S1; Additional file 1: Figure S1).
Second, molluskan group shows a wider range of distribution for theoretical isoelectric point (pI) values across MT isoforms as compared to mammalian and teleostean MT groups (Additional file 1: Figure S1). In mammals, MT-IIIs are only the isoform family (but excluding murine MT-IIIs) to show a significantly acidic pI values (4.79~4.82), while most other isoforms belonging to MT-I, −II or -IV class typically represent pI values higher than at least 7.5. Most teleostean MTs show pI values ranging from 7.8 to 8.4. On the other hand, bivalve MTs display pI values ranging 4.31 to 8.56 depending on isoforms. In the Gastropoda group, the pI values of the MT proteins range from 5.16 to 8.28. Although the numbers of taxa available for MTs from Gastropoda class were currently limited, theoretical pI values estimated from the category group Vetigastropoda (pI 5.16~6.00) were relatively lower than those from Heterobranchia (pI 6.10~8.28). Possibly, MT proteins with largely different pI values might exhibit differential capability to bind or interact with charged molecules in the host protective process. In addition, contents of some positively charged aa (e.g., Lys) may influence importantly the metal-binding function of MT protein. These aa residues (particularly at positions in vicinity to Cys) are thought to be involved in the stabilization of the interaction between MTs and metal ions through the electrostatic interactions to bridge the protonated basic residues and the negatively charged metal-thiolate complex (Pedersen et al. 1994). Thereby, such a wide range of pI values may be potentially suggestive of, at least in part, dynamic diversification and subfunctionalization among molluskan MT isoforms, although the biological and/or functional implications of such a wide range of pI values should be explored in the future.
Third, molluskan taxa are found to have a tendency of relatively lower Cys content (mean ± s.d. = 28.1 ± 1.8%) in their MT proteins than are mammalian (32.4 ± 1.3%) or teleostean (33.3 ± 0.3%) groups. Also, the intra- and interspecies variations in Cys contents of MTs are larger in mollusks (especially in bivalves; ranging 21.2 ~ 31.9%) than in mammals (26.2~34.4%; 31.1~34.4% if MT-IIIs excluded) and fish (32.8~35.0%) (Additional file 1: Figure S1). Metal binding property and capacity of molluskan MTs have been studied in only a few species (Palacios et al. 2011). However, Cys residues are known to be essential for the affinity to bind metal ions where the number of metal ions bound by the MT may be fundamentally determined by the number of Cys residues (Amiard et al. 2006; Jenny et al. 2016; Vergani et al. 2005). Hence, it is able to suggest that molluskan MTs may display relatively larger variations in the metal binding capacity among isoforms than vertebrate orthologue groups.
Taken together, all the three parameters above mentioned are undoubtedly indicative of large structural variations and divergence of MT proteins among molluskan species. Based on this overview, taxa (or lineage)-specific patterns for structural diversification of MTs are described more details in following sections, with a particular attention on the arrangement of Cys motifs.
Structural diversity of MT isoforms in major molluskan taxa
Nomenclature and classification of MTs used in this paper were referred to GenBank (NCBI) based on the definition of each sequence. If the MT sequence was published in scientific paper(s), its classification was checked again. Within a given species, the redundant MT sequences at aa level were not included in analyses (Additional file 1: Table S1). Classification of putative domain structure in each MT sequence was based on the number and arrangement pattern of Cys motifs as described previously (Jenny et al. 2016), since there have been no empirical studies on three-dimensional structures of molluskan MT proteins. In general, domain structure in MT is designated α and β (Braun et al. 1986; Binz and Kagi 1999). Usually, the α-domain contains eleven to twelve Cys residues, binds four divalent metal ions and confers structural stability on the MT polypeptides. On the other hand, the β-domain, binds three divalent metal cations through the nine Cys residues and participates in metal exchange reactions via glutathione-shuttling with metal-requiring apoproteins (Jiang et al. 1998; Jiang et al. 2000).
Gastropoda MTs
In a total, nineteen non-redundant MTs (from twelve species belonging to seven families) including a putative MT predicted in the unplaced genomic scaffold sequence (Biomphalaria glabrata; Planorbidae) were analyzed with sequence alignments. Among the nineteen sequences, 17 sequences with 59 to 70 aa residues are found to be fairly aligned in the multiple sequence alignment trials. In spite of substantial differences in non-Cys residues among taxa, they share a conserved pattern of Cys motifs. Eighteen Cys in these gastropoda MTs are arranged as [Cys-X-X-X-Cys → (Cys-X-Cys)3 → Cys] → [(Cys-X-Cys)2 → Cys → (Cys-X-Cys)2] (Additional file 1: Figure S2). It indicates that gastropoda MTs represent the protein structure comprising of two distinguished β-domain forms (i.e., β2β1-form) designated by the recent suggestion to propose the presence of two hypothetical ancestral β-domains (Jenny et al. 2016). The β2-domain structure at the N-terminal of gastropod MT protein is similarly observed in C. virginica MT-IIIs as well as in the β-domains of vertebrate (mammals and fish) MTs. On the other hand, the C-terminal β1-domain of the gastropod MT is commonly found in various molluskan MTs. According to the β2β1-structural scheme, the shortest P. acuta MT (comprised by 59 aa) is thought to have lost two Cys in its β2-domain. In that alignment, MTs from Vetigastropoda species possess more aa residues (5 or 8 aa) in the intervening region between the β2- and β1-domains than those from Heterobranchia species (2 aa).
Besides the common β2β1-shape, gastropod group represents two significantly lengthy MT polypeptides. One is 100-aa MT from Littorina littorea (Caenogastropoda; AY034179) (English and Storey 2003) and the other is 124-aa MT (XP_013080485; deduced from the unplaced genomic scaffold) from B. glabrata (Additional file 1: Figure S2). Based on manual alignment, L. littorea MT is proven to show a conserved pattern for Cys arrangements in N- and C-terminal parts (i.e., nine Cys respectively in putative N- and C-terminal domain regions). Considering the Cys motif patterns, the N- and C-terminal parts of the L. littorea MT could be designated β2- and β1-domains, respectively. Further, a closer examination on the intervening region (comprised by 32 aa) between the β2- (N-terminal) and β1- (C-terminal) domains has indicated that the 32-aa internal segment has been potentially a duplicated copy of the N-terminal β2-domain. It conserves clearly 9 Cys residues and shows a considerably high sequence identity (75%) to the N-terminal (β2-domain) domain. Based on our peer review, the structure of L. littorea MT could be considered as a novel shape of gastropod MT characterized by β2β2β1-domain form. Hence, this newly proposed structure suggests that domain duplication event might have served as a driving force to figure the large MT in certain gastropod taxa.
Another example for the domain duplication in gastropod MT is the 124-aa B. glabrata MT (XP_013080485). In that MT polypeptide sequence, three putative domain regions sharing a considerable sequence similarity in one another could be identified, and each of the three putative domains may be designated β2-structure based on their Cys arrangement patterns (Additional file 1: Figure S2). As numbered from the N-terminal, the first β2-domain and the second β2-domain share the conserved Cys motif frame (except one additional Cys in the second β-domain). They also show the high sequence homology (76.5%) each other. The putative third β2-domain linked to the second β2-domain is found to display 66.7% of sequence homology to the first and second β2-domains. Within this context, the B. glabrata MT could be proposed to possess at least three duplicated β2-domains tandemly arrayed in a tail-to-head fashion. On the other hand, the remaining C-terminal part (23-aa) following the β2β2β2-domain region is found to contain five Cys (three singlet Cys and a Cys-Cys doublet motif). Unlike the L. littorea MT above, the C-terminal region of this B. glabrata MT display no typical shape to be categorized into one of known domain structures (Additional file 1: Figure S2). Currently, the origin of this C-terminal region has been unknown. Further validation of scaffold genomic sequences along with mining of similarly organized MTs from other Heterobranchia genomes would be needed to get a deeper insight into the mechanism responsible for the formation of the array of three β2-domain region in this gastropod MT. This pulmonate snail species has already been known to express functionally diversified (i.e., different metal selectiveness) MT isoforms [(i.e., Cd-MT (GQ205374), Cu-MT (GQ205373) and intermediate Cd/Cu-MT (GQ205375)] (Berger et al. 1997). Hence, it would also be valuable to examine the expression patterns of this newly identified MT regarding its potential differentiation in physiological function and/or metal responsiveness in comparison with previously characterized paralogs from this species.
Bivalve-ostreidae MTs
Structural diversity of MT families in Ostreidae has been comprehensively described with C. virginica model (Jenny et al. 2004; Jenny et al. 2006). Currently, seventeen C. virginica MT sequences available in GenBank could be classified into one of four MT isoform families (MT-I, −II, −III, and IV). The MT families consist of 2 (MT-IA and MT-IB), 8 (MT-IIA, MT-IIB, MT-IIC, MT-IID, MT-IIE, MT-IIF, MT-IIG, and MT-IIH), 3 (MT-IIIA, MT-IIIB, and MT-IIIC), and 3 (MT-IVA, MT-IVB, and MT-IVC) subisoforms, respectively. In addition to these 16 sequences, one MT sequence named MTA (GenBank accession no. AF506977) is independently recorded in this oyster species (see Additional file 1: Table S1). Of the 16 isoforms, the prototypical MT structure (corresponding to MT-I form; MT-IA and MT-IB) is 75-aa MT possessing 21 Cys residues (28% of Cys content). The MTA isoform can also be classified to the same prototype (i.e., MT-I) with an addition of Asp in the region between 16th and 17th Cys residues. They share a high sequence homology one another in a prototypic αβ1-domain-structure (Jenny et al. 2004). This αβ1-structure is well conserved also in other oyster species, including C. gigas (Pacific oyster; AJ242657), C. ariakensis (the Suminoe or Asian oyster; DQ342281) and C. rivularis (the Jinjiang oyster; JN225502). The same domain structure is also relevant with isoforms from non-Crassostrea species Ostrea edulis (Tanguy et al. 2003). However, relative large substitutions of non-Cys aa (also the replacement of three Cys with other aa in the O. edulis MTb isoform) have been found in the O. edulis MTs compared to MT-I orthologs from Crassostrea species (Additional file 1: Figure S3A).
C. virginica MT-II family includes eight subisoforms (MT-IIA to MT-IIH), and this MT-II family has been known to be classified into two subgroups. First, the MT-IIA/-IIB group possesses a sole α-domain of which sequence is highly conserved with that of the prototypic αβ1-MT-I. The loss of functional β1-domain has been proposed to occur due to the point mutation in the linker region (i.e., non-sense mutation resulting in a stop codon). Consequently, MTs belonging to this group reveal noticeably short polypeptide length (43 aa) (Jenny et al. 2004). Second, on the contrary, the remaining six C. virginica MT-II isoforms are lengthy polypeptides (94-aa MT-IIC, 149-aa MT-IID/-IIE, 145-aa MT-IIF, 204-aa MT-IIG, and 200-aa MT-IIH). They have tandemly duplicated copies of α-domain where the numbers of repetitive duplications are variable among isoforms. In MT-IIC, only one duplication event is predicted (i.e., two tandem duplicate copies), while subisoforms MT-IID/-IIE/-IIF and MT-IIG/-IIH display tandem arrays comprised of three and four α-domain copies, respectively (Jenny et al. 2004). Between and among duplicated (repetitive) copies, a few aa substitutions have been found. Collectively, the divergent process of C. virginica MT-II family has occurred through the loss of β1-domain (e.g., MT-IIA/-IIB; α-domain-structure) followed by further divergence into various subisoforms having differential numbers of duplicated α-domains (e.g., MT-IIC to MT-IIH; α (n = 2 ~ 4)-domain structure) (Jenny et al. 2016).
However, this divergence pattern has not been always a common finding in the Ostreidae lineage (Additional file 1: Figure S3A). Rather than the loss of β-domain, C. gigas MT-II family has represented the tandem duplication of 32-aa β1-domain with retaining the α-domain, giving rise to the αβ1β1-domain structure (Tanguy and Moraga 2001). Further, unlike in C. virginica, there has been no variation in repeat numbers among C. gigas MT-II subisoforms. MT (AF349907) from another Crassostrea species, Portuguese oyster (C. angulata) has exhibited the duplication of only a short, partial β1-domain fragment (7-aa region), giving rise to a non-canonical αβ1β1P-structure with truncated C-terminus. Because there has been no other publically released MT paralog from C. angulata, it has been yet unclear if this oyster species may possess any paralog copies representing the complete αβ1β1-domain structure or not. Beyond the Crassostrea genus, our survey against GenBank has identified that Alectryonella plicatula MT (KP875559; 107-aa) should also display the typical αβ1β1-domain structure. Moreover, the A. plicatula MT shows very high sequence homology to its Crassostrea orthologs (MT-IIs), indicating the common origin of this multi-domain structure. Currently, the known αβ1β1-structured MT subisoforms in Ostreidae (except for the C. angulata MT with a truncated C-terminus) share the same N-terminal (Met-Ser-Asp-Pro) and C-terminal (Cys-Lys-Lys) motif residues (Additional file 1: Figure S3A).
On the other hand, the C. virginica MT-III group consists of three homogenous subisoforms MT-IIIA/-IIIB/-IIIC. They share each other high sequence identity including 18 Cys residues, and the distribution pattern of the 18 Cys have been proposed as the array of two β-domains as [(Cys-X-Cys)4 → Cys] × 2 (i.e., β2β2-MT) (Additional file 1: Figure S3B). The arrangement pattern of nine Cys is obviously similar with the β2-domain of the gastropod β1β2-MTs (Jenny et al. 2006; Jenny et al. 2016). The same β2β2-domain structure has also been found in C. gigas MT-III (JF781299); however unlikely in C. virginica, multiple MT-III subisoforms have not been characterized in C. gigas (Cong et al. 2012). The C. gigas MT-III shows a series of substitutions of non-Cys aa from the C. virginica MT-III isoforms. The overall sequence identity of MT-IIIs between the two Crassostrea species is about 70% (Additional file 1: Figure S3B). Meanwhile, Crassostrea MT-III isoforms reveal considerably low pI values (4.38~4.77 for C. virginica and 4.31 for C. gigas) as compared to other MT isoforms showing pI values > 7.5. With the viewpoint of the low pI value, C. virginica MT-III may resemble mammalian MT-III family. Most known mammalian MT-III isoforms except murine MT-IIIs reveal acidic pI ranges (4.79~4.82) with acidic 6-amino-acid insert in the C-terminal region. Synthesis of mammalian MT-III is not inducible by heavy metals and localized predominantly in the central nervous system (Faller 2010). Unique roles of mammalian MT-III differing from other MT isoforms have been characterized as the neuronal growth-inhibitory factor to inhibit neuronal outgrowth (Wang et al. 2006). Specific roles of bivalve MT-III differing other MT family groups have not been yet extensively addressed. However, the expression study with C. virginica MT-III has indicated that the C. virginica MT-III showed quite a low basal level of expression in adult tissues (i.e., only actively expressed in early larvae). Further, C. virginica MT-III represented only a moderate responsiveness to heavy metal exposures in both larvae and adults (Jenny et al. 2006). However, on the contrary, the C. gigas MT-III has been reported to be significantly induced by zinc (as a main regulator for zinc homeostasis), and it may be a participating member for cadmium detoxification in the adult tissues (Cong et al. 2012). Thereby, it suggests the functional differentiation/divergence of MT-III isoforms during speciation events in the Crassostrea lineage. Within the context of this hypothesis, it is worthy to have an attention on another C. gigas MT isoform named mt3 under the accession number AJ295157. This C. gigas mt3 isoform is not a true MT-III family member with the β2β2-domain structure. Rather than, mt3 should be considered as a MT-I member because it represents the αβ1-domain structure (see Additional file 1: Figure S3A). However, due to the significant non-Cys aa substitutions, C. gigas mt3 displays only 67% sequence identity to its paralogue MT-I isoform. Moreover, the substitutions include the change of three Lys residues to uncharged aa as well as the replacement of uncharged aa with negatively charged aa. Such aa substitutions might give rise to lower pI value (5.98) of mt3 than those of its paralogue MT-I members. A previous study has reported that mt3 should have the extremely low basal expression level with only moderate or minute responsiveness to metal exposure (Marie et al. 2006). The mt3 has been suggested to have probably no significant physiological functions under metal exposure and to be expressed only in particular developmental stages (Marie et al. 2006). Hence, taken together, it could be hypothesized that C. gigas mt3 might have been diverged from the prototypic MT-I through the non-Cys aa changes. This isoform is likely to show, at least in part, certain functional orthology to the C. virginica MT-III, although they have different domain structures.
Finally, the MT-IV isoforms (MT-IVA, MT-IVB and MT-IVC; 83 aa in length) from C. virginica have been proposed as variant forms of αβ1-MT. This isoform group has been supposed to have experienced a series of aa substitutions including Cys residues, giving rise to 25 Cys with the formation of a Cys-Cys doublet (in the α-domain) and three Cys-Cys-Cys triplet motifs (in the β1-domain). The C-terminal residue of the C. virginica MT-IV isoform (glutamine-alanine-threonine) is also notably different from those of other paralog isoforms. Thereby, the proposed designation of domain structure for MT-IVs could be α′β1′-form. In addition to C. virginica, two Crassostrea species (C. gigas and C. ariakensis) possess MT-IVs of which primary domain structures are fairly conserved with that of C. virginica MT-IV. However, C. gigas MT-IV (87 aa; AM265551) and C. ariakensis MT-IV (86 aa; JF919323) represents one additional Cys residue at C-terminal region (Additional file 1: Figure S3B). Besides the above C. gigas MT-IV clone (AM265551), genome sequencing of C. gigas (Zhang et al. 2012) represents two unplaced genomic scaffolds [scaffold852 (JH816574) and scaffold1297 (JH818394)] each containing the putative gene encoding MT (EKC32371 and EKC28510, respectively, in the two scaffolds). The deduced sequences of these MTs are 128 and 137 aa in length, respectively. Both MTs are predicted to possess unusual N-terminal regions (41-aa for EKC32371 and 50-aa for EKC28510). However, immediately following the N-terminal region, the two putative MTs represent the 87-aa-structure that is apparently homologous to the C. gigas MT-IV isoform (AM265551). When the unusual N-terminal regions are excluded from these two MT-IV-like sequences, the three MT sequences (one characterized MT-IV and two in-scaffold sequences) reveal only one aa substitution, although the scaffold sequences should be further validated in future.
Bivalve-mytilidae MTs
In Mytilidae, 33 full-length, non-redundant MT aa sequences were retrieved from five taxa (12 sequences form Mytilus edulis, 6 from M. galloprovincialis, 2 from Mytilus sp., 11 from Perna viridis and 2 from Bathymodiolus azoricus). Mytilid MTs are a structurally more homogeneous group as compared to Ostrea MT group. They represent 66-aa to 75-aa polypeptides containing 19 to 23 Cys residues, and display typically the αβ1-domain structure (Additional file 1: Figure S4).
In the mytilid mussels, two types of MT isoforms, MT10 and MT20 have been described previously (Aceto et al. 2011; Leung et al. 2014). These two MT types differ in mass and Cys arrangement. The monomeric form MT10 (~10 kDa) represents generally 73-aa polypeptides including 21 Cys residues mainly arranged as nine Cys-X-Cys motifs. On the other hand, the dimeric form MT20 (~20 kDa) is typically 72-aa polypeptides containing 23 Cys residues. Unlike MT10, the MT20 isoforms show a Cys-Cys doublet in the α-domain. MT10 and MT20 have been reported to be functionally differentiated. MT10 has been found to be more abundant than MT20, and hence it could be considered as a main player for the regulation of homeostasis under basal conditions (Leung et al. 2014; Lemoine et al. 2000). Under metal-exposed conditions, MT10 and MT20 have been known to display differential responses and binding ability to essential and non-essential metals. MT10 could be actively inducible by various metals while MT20 be more preferentially associated with non-essential metals such as Cd and/or Hg (Raspor et al. 2004; Dondero et al. 2005; Vergani et al. 2007).
From multiple sequence alignment, mytilid MTs could be categorized into three main groups (Additional file 1: Figure S4). The first group consists of three MT10B sequences (two from M. edulis and one from M. galloprovincialis; AJ577126, AJ577127 and DQ848984, respectively). They represent 66-aa polypeptides containing 19 Cys residues with the deletion of one Cys-X-Cys motif in the α-domain. All the three MT sequences are originated from intronless MT genes (Leignel et al. 2005; Yang et al. 2014). The second group contains twenty-four MT10 isoform sequences. They reveal 72 to 75 aa in lengths and possess 21 Cys residues in conserved positions (Lemoine et al. 2000; Khoo and Patel 1999). There are only two exceptions; one is the replacement of a Cys with Arg in P. viridis MT-IA (JN596471) and the other is an insertion of an additional Cys in P. viridis MT (AF036904). Within the second group, the MTs are likely to be sub-grouped according to known taxonomic appraisal at genus level (i.e., Bathymodiolus, Mytilus and Perna). All the P. viridis MT-II isoforms (named MT-IIA, −IIB, −IIC and -IID) are found to possess 72 aa, as similarly with the mytilid MT20 isoforms. Nevertheless, based on their Cys motifs frame, these P. viridis MT-IIs (JN596477 to JN596480) have been proposed as MT10 members. Previous phylogenetic analysis has claimed an early divergence of P. viridis MTs from the main mytilid MT10/MT20 groups (Leung et al. 2014). When aligned with other mytilid orthologs, P. viridis MT10-I and/or MT10-II represent several residues distinct from other mytilid MT isoforms. They include positions 11th (alignment position; Gln/Lys in P. viridis MTs vs. Asn in all other mytilid MT10 and MT20 isoforms), 62nd (Gln vs. Gly/Asp) and 74th (Ser vs. Gly). Further, P. viridis MT10 isoforms (both MT10-I and MT10-II) are found to share the same aa in several positions with mytilid MT20 isoforms. These could be exemplified by positions 34th (alignment position; Ser in P. viridis MTs and MT20s vs. Gly in other mytilid MT10s), 39th (Gly vs. Lys), and 73rd (Ser vs. Pro). Finally, the third group is comprised of six MT sequences from three Mytilus species. They are 72-aa polypeptide containing 23 Cys residues (i.e., MT20 type). Only one exception is the substitution of Cys to Arg in the M. edulis MT-20 clone. From the alignment, two MT20-specific residues could be found at positions 24th (Lys in MT20s vs. Glu in all MT10s) and 68th (Asn vs. Thr) (Additional file 1: Figure S4). In particular, the change from negatively charged aa (e.g., Glu) to Lys at position 24th is likely related with the MT20-specific formation of Cys-Cys doublet motif, since the positively charged aa (e.g., Lys) at vicinity to Cys motif is considered to play important roles in the stabilization of the metal binding reaction in most MT proteins (Pedersen et al. 1994).
Bivalve-other taxa
Besides the two main bivalve taxa (Ostreidae and Mytilidae), 27 non-redundant, full-length MT isoforms have been exploited from 21 bivalve species belonging to one of six orders Pterioida, Arcoida and Pectinoida (subclass Pteriomorphia), Pholadomyoida (subclass Anomalodesmata), Veneroida (subclass Heteroconchia), and Unionoida (belonging to Palaeoheterodonta). From the multiple sequence alignment, most of them are found to represent αβ1-domain-structure with a conserved 21-Cys-frame (Additional file 1: Figure S5A). However, several variant isoforms are also found to show modification(s) of Cys motifs in one or two positions, giving rise to the generation of Cys-Cys doublet, substitution, insertion or deletion. Several variant isoforms are found to retain the total number of Cys residues (i.e., 21 Cys) while others show changes of the total number of Cys residues. MT isoforms from Pinctada maxima (pearl oyster; FJ389580) (Tang et al. 2009) and Laternula elliptica (Atlantic clam; DQ832722/DQ832723) display the insertion of an additional Cys in the third Cys-X-Cys motif, resulting in the Cys-Cys-Cys motif at that position. On the contrary, an MT isoform from Hyriopsis schlegelii (freshwater pearl mussel; MT2; KJ019821) has lost one Cys residue at the second Cys-X-Cys motif along with considerable alterations in non-Cys aa residues. Unlike its paralog (H. schlegelii MT1; KJ019820), the H. schlegelii MT2 has been proposed as a genetically separated isoform (Wang et al. 2016). A recent study has indicated that these two H. schlegelii MT paralogs might have been subfunctionalized as evidenced by clearly distinct tissue expression patterns (i.e., constitutive expression of H. schlegelii MT1 vs. gonad-specific or predominant expression of H. schlegelii MT2) (Wang et al. 2016). Another example for the large difference between paralog MT isoforms is P. martensi MT isoforms. Even though the P. martensi MT1 (KC197172.1) represents the common αβ1-shape, its paralog MT2 (KC832833.1) exhibits an apparently non-canonical pattern of Cys arrangement (20 Cys).
On the other hand, two MT isoforms from Veneroida are found to have noticeably less number of Cys residues than others: one is the duck clam Mactra veneriformis (Mactridae) MT with 18 Cys (59-aa; Cys content = 30.5%; FJ611963) (Fang et al. 2010; Fang et al. 2013) and the other is Venus clam Cyclina sinensis (Veneridae) MT with 16 Cys (74-aa; Cys content = 21.6%; HM246244) (Lü et al. 2012). The M. veneriformis MT seems to have lost an internal fragment near N-terminal region (possibly corresponding to the α-domain) containing three Cys residues (likely a Cys-X-Cys motif and one conserved Cys). The C. sinensis MT lacks a Cys-X-Cys motif in α-domain and additionally three Cys residues (Cys-X-Cys motif and one Cys residue) probably in the β-domain.
Importantly, three MT isoforms display large polypeptide sizes comprising of more than two putative domains (Additional file 1: Figure S5B). Of the three MTs, two MT sequences (MT1 and MT2) are from the bay scallop Argopecten irradians (Pectinidae) and remaining one isoform is from the fingernail clam Pisidium coreanum (Sphaeriidae). These sequences have been reported earlier but their domain structures have never been addressed clearly. Even though A. irradians MT1 (145-aa; 40 Cys; EF093795; (Liu et al. 2006)) and MT2 (110-aa; 28 Cys; EU734181; (Wang et al. 2009)) exhibit essential features of mollusk MTs (i.e., the presence of characteristic Cys-X-Cys motifs), their overall structures are more or less complicated and difficult to be simply categorized into one of currently known shapes of bivalve MTs. However, in a broad sense, these isoforms may bear a resemblance to the MT isoforms with multi-β-domain-structure. For both A. irradians MT isoforms, the C-terminal region may be considered the β1-domain possessing nine Cys as (Cys-X-Cys)2 → Cys → (Cys-X-Cys)2. In addition to the C-terminal β1-domain, A. irradians MT1 potentially exhibits three tandemly arrayed β2-like domains. However, each β2-like domain in the A. irradians MT1 displays some non-canonical arrangement of Cys. First, two Cys of the first Cys-X-Cys motif in each domain is separated further by intervening 2~4 aa residues (i.e., similar with the pattern found in N-terminal regions of gastropod β2-domains). Second, Cys-Cys doublet motifs rather than a canonical Cys-X-Cys motif are present in the first and third β2-like domains. Third, an additional Cys-X-Cys motif exists in the flanking regions between the first and second β2-like domains as well as between the second and third β2-like domains. Nevertheless, the overall shape of A. irradians MT1 may be designated β2β2β2β1-like structure, although this novel proposal should be further challenged with empirical structural analyses. Sequence comparisons among/between these successive β2-like domains indicate that they share little sequence similarity one another except conserved Cys residues (Additional file 1: Figure S5B). Within this scheme, the A. irradians MT2 could be treated as a paralog having the less number of β2-domains (i.e., putatively designated β2β2β1-like structure). The A. irradians MT2 also reveals some non-canonical attributes including the lack of one Cys residue and the formation of Cys-Cys doublet in the N-terminal β2-like domain. These two A. irradians MT isoforms are found to share only a little sequence homology, indicating that they may be quite distantly related paralogs (Wang et al. 2009).
On the other hand, the 105-aa P. coreanum (Sphaeriidae) MT (GQ268325; 31 Cys) (Baek et al. 2009) is found to show the domain multiplication to resemble the αβ1β1-structure, of which Cys arrangement is similar with that of C. gigas MT-II. However, unlike C. gigas MT-II to show the tandem array of two homogenous β1-domains, P. coreanum MT contains the two heterogeneous β1-domains with no apparent sequence homology between the two domains. The P. coreanum MT lacks a common triplet linker sequence (Lys-Val-Lys/Val) between α- and first β1-domain. The array of two heterogeneous β1-domains linked to N-terminal α-domain observed in P. coreanum MT could be the novel structure of bivalve MT proteins (Additional file 1: Figure S5B).
Domain evolution in molluskan MTs
Currently, the proposed hypothesis for the evolution of molluskan MT has been based on the domain duplication event(s) from an ancestral single domain-structured MT, in which the β-domain has been considered as the ancestral shape (Cols et al. 1999). After an early duplication event of the ancestral β-domain, the resultant ββ-domain MT has undergone divergent processes, given rise to the αβ-structure in certain taxa (Jenny et al. 2016; Braun et al. 1986; Cols et al. 1999). Difference in the metal-binding properties between the α-domain and the β-domain makes the two domains to represent differentiated roles in the cellular physiology. Generally, α-domain plays a more prevalent role in Zn homeostasis and detoxifying sequestration of toxic metals (e.g., Cd) whereas the β-domain is primarily responsible for the homeostatic regulation of essential metals (e.g., Cu) (Jenny et al. 2004; Cols et al. 1999; Nielson and Winge 1984; Xiong et al. 1998). Consequently, the multi-domain MT in specific taxa acquiring both α- and β-domains was able to perform the dual functions; the detoxification of toxic metals by the α-domain and the homeostasis of physiologically relevant metals by the β-domain (Jenny et al. 2016; Cols et al. 1999; Nielson and Winge 1984; Nielson and Winge 1983).
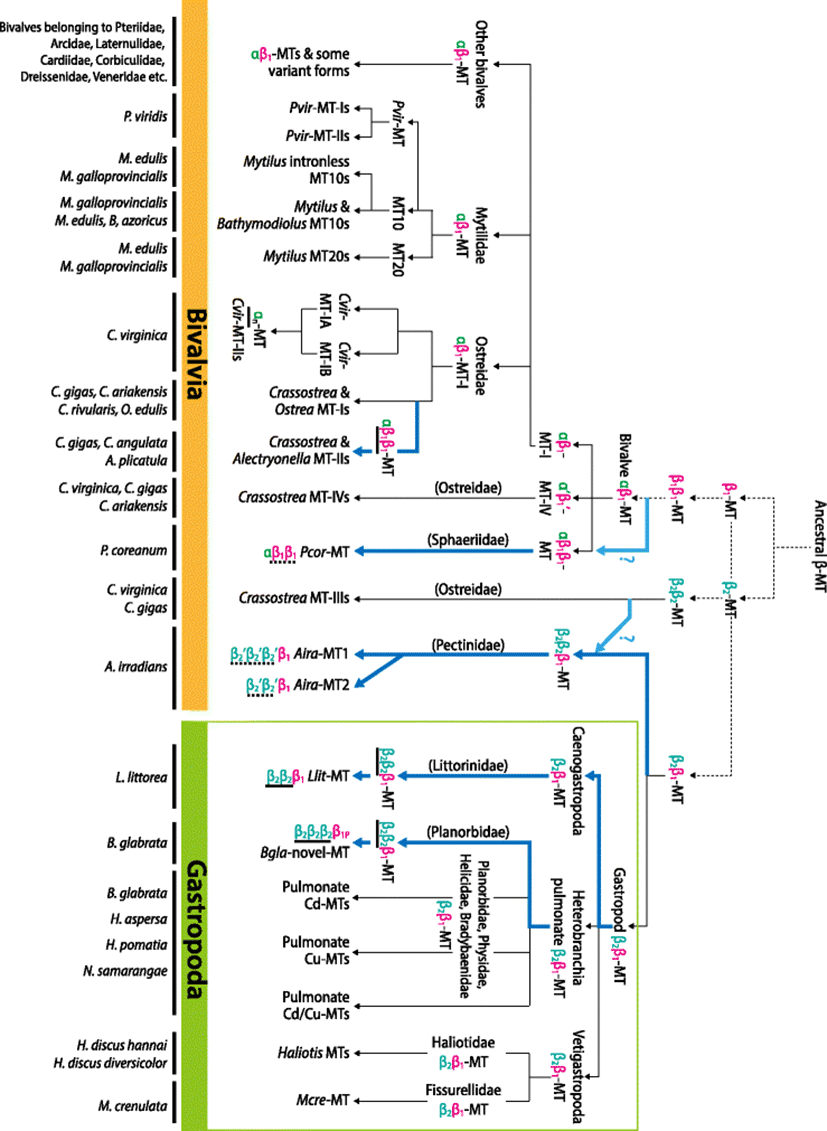
The latest phylogenetic work has proposed that two distinct ancestral β-domains (designated β1 and β2 domains) might have existed and given rise to the structural diversity of all molluskan MTs (Jenny et al. 2016). In that literature, they have hypothesized separate paths of the evolution of the two ancestral MTs in the major taxa within the mollusk phylum. The two β-domains appear to have diverged into two structurally different MT isoform types (i.e., αβ1-MT and the β2β2-MT) in bivalves whereas in gastropods, the two ancestral β-domains form a single structural β2β1-MT isoform. With C. virginica model, the structural diversity of bivalve MT isoforms has been highlighted to demonstrate evolutionary paths from not only αβ1-domain but also β2β2-domain (Jenny et al. 2016). On the contrary, in the gastropoda lineage, the β2β1-domain structure has been proposed as a typical appearance common to most extant gastropod MTs. Instead of a series of domain duplications seen in bivalve MTs, gastropod MTs appears to have diverged to functionally differentiated isoforms (i.e., Cd-MT, Cu-MT or intermediate Cd/Cu-MT) through the composition changes of non-Cys aa residues (Jenny et al. 2016; Palacios et al. 2011; Cols et al. 1999). However, our bioinformatic analyses in this study suggest that the current theory on MT domain evolution in the phylum Mollusca could be revised based on newly recognized evidences. Novel hypothetical paths and additional insights into the domain evolution of molluskan MTs are proposed in following sections.
The non-canonical domain structure of large MTs from two gastropod species (L. littorea and B. glabrata) may be considered as novel shapes. Phylogenetic analysis of gastropoda MT domains (β1 or β2) has generated the two major clades separated depending on the types of β-domains (β1 or β2) (Fig. 1). The gastropod β2-clade has been proven to contain all previously proposed gastropoda β2-domains together with the putative β2-domains of the large MTs proposed in this study. For the large B. glabrata MT, three β-domains (the first to third domains numbered from the N-terminal) are closely clustered together and placed in the major clade comprising the gastropod β2-domains. This result suggests obviously that they are duplicated copies of β2-domains that might have evolved through the tandem duplication events. On the other hand, the C-terminal region containing only five Cys is not clustered with any typically known β1 or β2-domain sequences. Although we did not provide clear evidence for the origin of this C-terminal region, the most likely scenario is that the originally existed β1-domain at C-terminal in the ancestral β2β1-MT might have undergone certain recombination(s) including the loss of some parts during duplication events of neighboring β2 domains. Based on this assumption, the unusual C-terminal part of B. glabrata MT might be a reminiscent, partial segment (designated β1P here) originated from the early β1-domain. Hence, the tandem duplications of β2-domain accompanied with partial loss of the C-terminal β1-domain may be a plausible mechanism to produce the current β2β2β2β1P structure in this pulmonate species. We performed additional analyses on the duplicated β2-domains of this large MT (first and second β2- domains used for analysis) in order to hypothesize a plausible reason responsible for the happening of this evolutionary episode. For this, β2-domains of previously known MTs from pulmonate species (i.e., Cd-MT, Cu-MT and intermediate Cd/Cu-MT) were included in analyses together with the β2-domains of this large B. glabrata MT. From the sequence alignment, the duplicated β2-domains of the B. glabrata β2β2β2β1P-MT revealed a more sequence similarity to Cd-MT than to Cu-MT and Cd/Cu-MT. Phylogenetic analysis of β2-domains from B. glabrata paralogs also showed a close relationship between the β2β2β2β1P-MT and Cd-MT (Additional file 1: Figure S6). Although the tree topology on the affiliation was not statistically supported, it could be enough to hypothesize that the emergence of β2β2β2β1P-MT in B. glabrata might have been an evolutionary process toward the need of more specificity for detoxification of non-essential metals (i.e., primarily Cd). This hypothesis is congruent with the evolutionary theory of multi-domain MTs in bivalves. In bivalves, development of Cd-preferring MT has been proposed to be based on the conversion of a Cu-preferring β-domain to the Cd-preferring α-domain by the acquisition of additional Cys, followed by subsequent domain duplications (Jenny et al. 2004; Jenny et al. 2006). On the other hand, in pulmonate gastropods, it has been widely proposed insofar that further domain duplication from the prototypic β2β1-MT form has unlikely happened. Instead, specific MT isoforms with different metal selectiveness in pulmonate gastropods have been achieved mainly through the composition changes of non-Cys aa (Jenny et al. 2016; Palacios et al. 2011). However, from the new evidence in this study, domain duplication giving rise to large MTs should be considered as one of the important mechanisms permitting pulmonate MTs to achieve more specificity for their cognate heavy metals. Taking into account that β2β2β2β1P-MT-originated β2-domains display much closer relationship among themselves than with the Cd-MT-originated β2-domain in the phylogenetic analysis, it is likely that the divergence to the β2β2β2β1P-MT in the B. glabrata genome might have occurred through a separate path independent of the pathway for generating the Cd-MT (Fig. 2). Hence, exposure experiments to examine metal selectiveness or binding property of β2β2β2β1P-MT domains would be helpful to test this hypothesis.
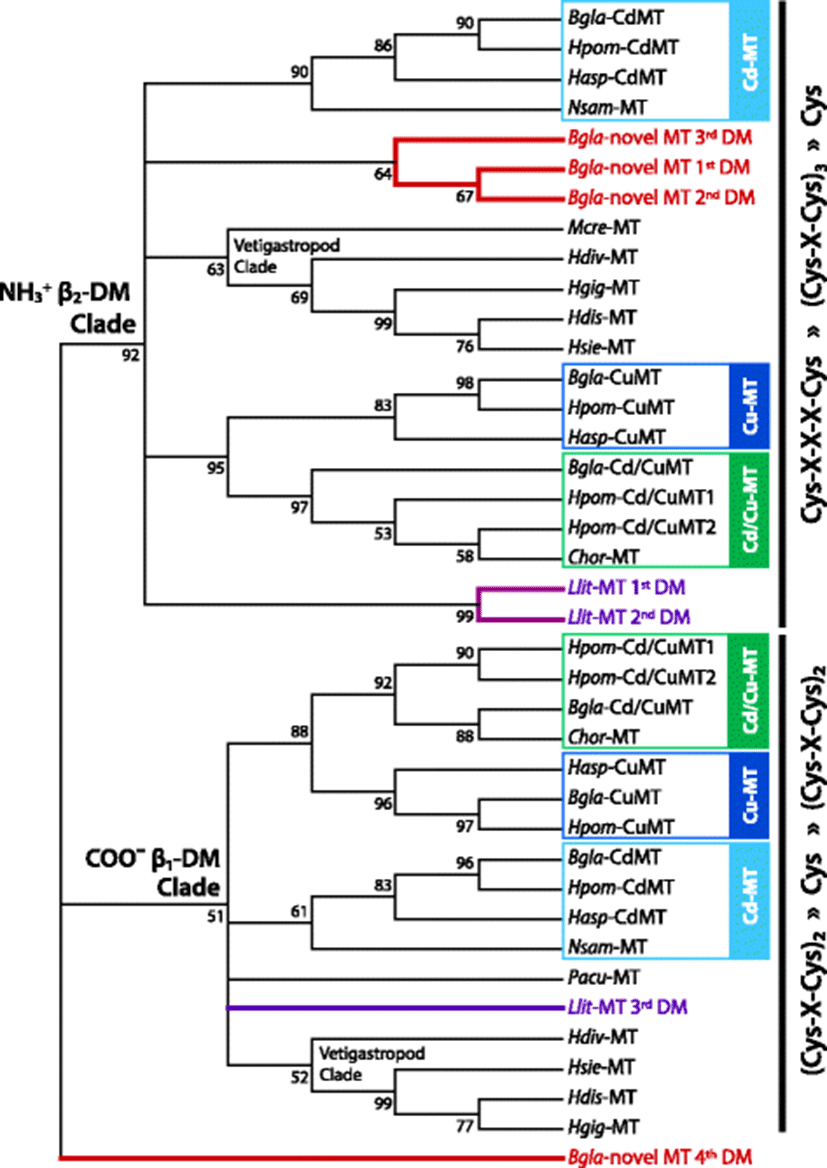
The other evidence for the β2-domain duplication in the gastropod MTs is observable in the periwinkle (L. littorea; Caenogastropoda; Hypsogastropoda) MT. In the molecular phylogenic tree, a subclade consisting of two closely affiliated L. littorea β-domains (first and second domains numbered from the N-terminal) was placed within the gastropod β2-clade, whereas the third β-domain of the L. littorea MT was positioned in the gastropod β1-clade (Fig. 1). Based on the phylogenetic separation between first, second, and third β-domains indicates that the L. littorea MT is comprised of two successive β2-domains from the N-terminal that is linked to the C-terminal β1-domain (i.e., β2β2β1-MT). Like the B. glabrata β2β2β2β1P-MT above, the first and second β2 domains in the L. littorea MT share high sequence similarity including the conserved Cys motifs, suggesting that they might have evolved from a tail-to-head tandem duplication event (Additional file 1: Figure S2). Further efforts to exploit potential paralog isoforms from this species or closely related species are needed to hypothesize potential factor(s) to drive the domain duplication in L. littorea MT.
Postmortem studies have claimed that most gastropod MTs would be very conservative in the Cys arrangements as a β2β1-domain structure (Jenny et al. 2016; Berger et al. 1997; Dallinger et al. 1993). On the contrary, substitutions/replacements of non-Cys aa residues while retaining the β2β1-frame have been thought as the major process for the evolutionary divergences of MTs in this primitive class Gastropoda. The Cu homeostatic requirements (thought to be mainly operated by β-domains) from the use of hemocyanin as a respiratory pigment in these gastropods, which is not present in oysters, has also been proposed as one of plausible factors responsible for the lack of divergently duplicated domains in gastropod MTs (Jenny et al. 2016; Berger et al. 1997; Perez-Rafael et al. 2012; Perez-Rafael et al. 1844). However, the present study claims that the formation of large MT with more than two domains should not be a bivalve-exclusive episode. It might have been an important path allowing some gastropod MTs to better modulate metal specificity in response to variations occurred in their habitat environments (Fig. 2). For both examples (B. glabrata MT and L. littorea MT), the target domain that has undergone duplication is the β2-domain. Hence, novel or specified functions (e.g., detoxification of non-essential metals) could be assigned to duplicated β2-domains while original and fundamental roles (e.g., Cu homeostatic regulation) are retained in the β1-domain. Extra metal-binding residues offered by duplicated β2-domains may also be potentially advantageous to strengthen further capacity of both metal reservation and resistance to excessive metal ions. Taken together, hypothesis for evolutionary mechanism to mold gastropod MTs should be revised taking into account the inclusion of this novel path featured by the duplication of β2-domain.
On the other hand, unlike in Heterobranchia and Caenogastropoda, the clear sign for taxa-specific domain multiplication has not been yet identified in Vetigastropoda species (abalone and limpet) (Lee and Nam 2016; Lieb 2003). Although the characterization of MT in this taxonomic group has been very limited, vetigastropod species have reportedly shown only a single MT isoform (i.e., β2β1-MT) within a given species (Fig. 2). Currently, it is unclear if vetigastropods possess functionally or structurally diverged paralogs. However, a recent study has reported that the abalone (Haliotis discus hannai) MT would be responsive to not only various heavy metals including Cu, Cd, and Zn but also non-metal stimulating stress treatments such as induced hypoxia, immune challenge and heat shock (Lee and Nam 2016; Guo et al. 2013). Based on this observation, the MT, at least in this vetigastropod species, is thought to have evolved to play readily multifunctional roles in diverse pathways involved in stress physiology.
The evolutionary theory of MT based on domain duplication has been the most comprehensively highlighted in the Crassostrea species, particularly in C. virginica. Two ancestral β-domains (i.e., β1 and β2) appear to have diverged to produce two different structural MT isoforms, i.e., αβ1-MT and β2β2-MT in the oysters belonging to Ostreidae (Jenny et al. 2004; Jenny et al. 2006). The ancestral β1-domain appears to have duplicated to produce a two-domain-structured MT that ultimately led to the evolution of the αβ1-structured MTs, which is observable in the Crassostrea species MT-Is and MT-IVs. On the other hand, the Crassostrea species MT-IIIs reveal the typical β2β2-domain structure, which might have been a descendant shape resulted from the duplication event of a single ancestral β2-domain (Fig. 2). In the reconstructed phylogenetic tree, Ostreidae β-domain sequences are placed on one of two main clades (i.e., either β1- or β2-domain clade), although several subclades within the β1-clade are not supported by high confidence values (Additional file 1: Figure S7). Crassostrea MT-IVs are closely related each other and distinguished from MT-I/-II isoforms within the β1-clade, suggesting the early divergence between MT-IV and MT-I/-II families from the prototypic αβ1-MT form. Also within the β1-clade, it is notable that the C. virginica MT isoforms have a tendency to be separately clustered from MTs from other Crassostrea species (such as C. gigas, C. ariakensis, C. rivularis, and C. angulata) as seen in both MT-I and MT-IV groups. It may suggest the divergence of these MT families during speciation in the genus Crassostrea to make C. virginica to be distinguished from other Crassostrea species (Jenny et al. 2016) (Additional file 1: Figure S7 and Additional file 1: Figure S8). The most apparent difference between C. virginica and other Crassostrea species is found in the MT-II family. C. virginica MT-IIs have been reported to the most recently evolved isoforms, which have formed through the loss of β-domain, giving rise to the sole α-domain structured MTs (Additional file 1: Figure S8). However, this divergence pattern in C. virginica is clearly contrasted by the duplication of β1-domain in C. gigas MT-IIs (i.e., αβ1β1-structured MT), suggesting the different evolutionary paths of the MT-II between the two closely related species belonging to the same genus (Jenny et al. 2004; Tanguy and Moraga 2001). Such a solely α-domain-based structure has been found only in C. virginica insofar, whereas αβ1β1-domain structure has also been observed similarly in other Crassostrea species and non-Crassostrea species (Additional file 1: Figure S7). Meanwhile, the β2-domain clade comprising of C. virginica and C. gigas MT-III isoforms shows a monophyletic topology, indicating the divergence from a common ancestral β2β2-MT origin. However, within the β2-clade, two subclades are characterized by the first and second β2-domains rather than by species. All the N-terminally present β2-domains are clustered together in a subclade while all the C-terminally present β2-domains in other subclade (see also (Jenny et al. 2016)). This finding may be indicative of that the first and second β2-domains of the Crassostrea MT-IIIs might appear to have originated before the separation of the two oyster species (Fig. 2).
Mytilidae species represent uniformly the αβ1-domain structure (known as the prototypic shape of bivalve MT). Some mytilid species have been reported to possess the intronless, relatively short MTs in their genomes. The presence of intronless MT genes has been proposed as the organisms’ strategic means for the efficient response to changes of cellular metal circumstances through the rapid transcription of MT genes (Leignel et al. 2005). Molecular phylogenetic analysis of Mytilidae MT domains generated trees consistently comprising of three main clades; clades for mytilid MT10s, P. viridis MTs and mytilid MT20s (Additional file 1: Figure S9). In congruent with the previous phylogenetic results using the entire MT polypeptide region, present phylogenetic analyses using separate domains also suggest the early divergence of P. viridis MTs from the other mytilid orthologs (Leung et al. 2014). In the Mytilidae lineage, a critical event before speciation is the divergence of MT10 and MT20 forms (Aceto et al. 2011) (Fig. 2). As compared to MT10, MT20s are characterized by the acquisition of additional Cys residues (i.e., a Cys-Cys doublet) in their α-domains (Leignel and Laulier 2006). It could be thought as a process to prepare the paralogue MT varieties with better execution in the detoxification of non-essential metals, since more Cys residues are generally taken into account for enhanced capability for sequestrating the toxic metals (i.e., metal tolerance). This hypothesis could be supported by the fact that MT20 would be more preferentially associated or exclusive reacted with non-essential metals than MT10 (Leung et al. 2014; Lemoine et al. 2000; Vergani et al. 2007). Conversely, in some environmental situations, the early αβ1-MT might have diverged into functionally differentiated isoforms in the Mytilidae: MT10 to execute primarily the homeostatic regulation of physiologically relevant metals and MT20 to function in the detoxification of trace metals (Fig. 2).
Exploitation of genetic determinants for MTs from other bivalve taxa has often showed the species (or lineage)-specific variations in MT structure. However, currently limited volume of knowledge on these MTs still hurdles to hypothesize the evolutionary mechanism of non-canonical MT forms in detail. Reconstruction of molecular phylogenetic trees in this study displays two main clades: one is a large clade comprising of β1-domains from various taxa and the other is a small clade consisting of five presumed β2-domain sequences deciphered from two A. irradians (Pectinidae) paralogue MTs (Fig. 3). Within a former β1-clade, paralogue isoforms from a given single species (e.g., R. philippinarum MT1/MT2 and L. elliptica MT10a/10b) formed subclades supported by high bootstrap values. Similarly, several subclades consisted of orthologs from closely related species belonging to the same genus ([e.g., MTs from genus Meretrix (Chang et al. 2007; Wang et al. 2010; Jiang et al. 2016) and genus Cerastoderma (Desclaux-Marchand et al. 2007; Ladhar-Chaabouni et al. 2009; Paul-Pont et al. 2012)). Collectively, it suggests that they might have evolved from recent divergence at species or genus levels. In contrast, some paralogue MT isoforms are distantly placed in the phylogenetic tree, although they are placed in the same β1-clade. Such a distant relationship is found in the genus Hyriopsis where MT1 and MT2 paralogs are not affiliated depending upon species. Although nomenclatures MT1 and MT2 are not established clearly in this two species, an isoform of H. cumingii MT (GQ184290) is closely related with H. schlegelii MT1 ortholog (KJ019820), rather than its paralogue isoform (FJ861993). This finding may indicate that the divergence between MT1 and MT2 might have occurred earlier than the speciation of the two Hyriopsis species (Yang et al. 2014; Wang et al. 2016).
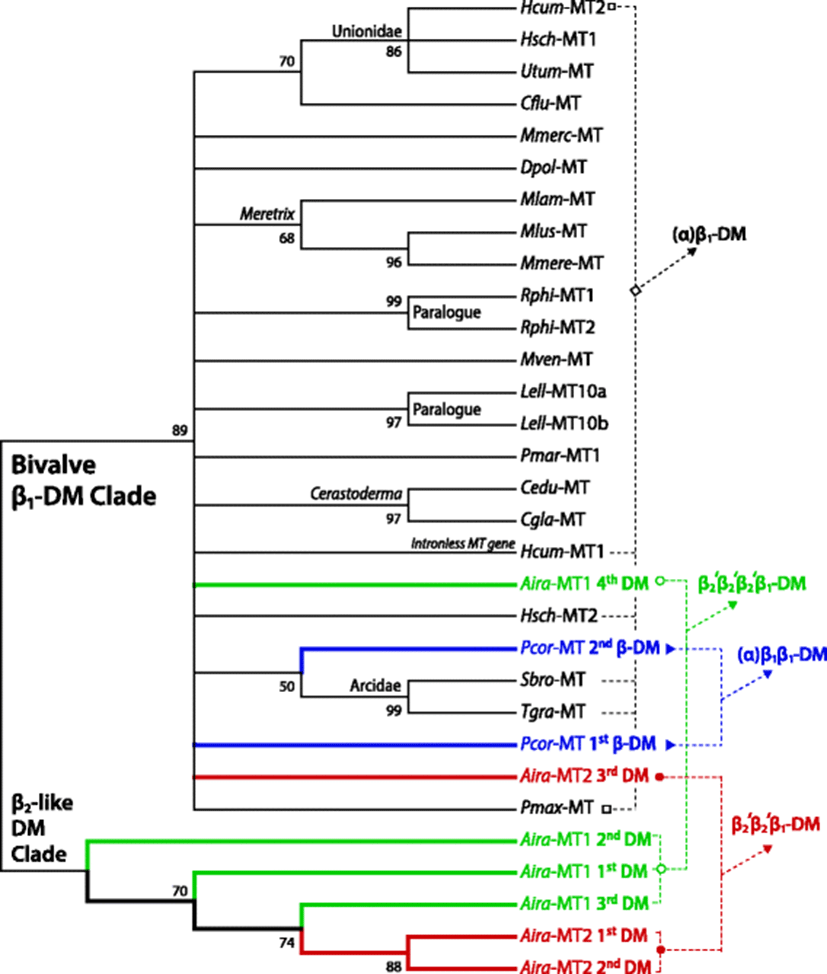
From the present molecular phylogenetic analysis, novel paths of MT evolution through domain duplication giving rise to large-sized MTs with more than two metal-binding domains could be proposed (Fig. 3). Evidences come from two bivalve species: one is P. coreanum (Sphaeriidae) (Baek et al. 2009) and the other is A. irradians (Pectinidae) (Liu et al. 2006; Wang et al. 2009). In the P. coreanum, the two putative β-domains are placed in the β1-clade. Within the β1-clade, the second (numbered from the N-terminal) β1-domain of P. coreanum MT is found to form a subclade with two β1-domains from Arcidae species [Scapharca broughtonii (FJ154101) and Tegillarca granosa (AY568678)]. Considering the N-terminally present putative α-domain, this P. coreanum MT could be designated αβ1β1-structure. In the phylum Mollusca, the αβ1β1-domain structure (i.e., duplication of β1-domain from the anticipated prototypic αβ1-MT) has been previously reported to be the Crassostrea-specific event (Tanguy and Moraga 2001). However, we have already proposed above that this event has also been true for non-Crassostrea oyster (i.e., A. plicatula; Ostreidae). Further, the present P. coreanum (Sphaeriidae) MT could indicate that this duplication process would not be limited to the Ostreidae. However, the evolutionary scheme for the domain duplication may be different between the two families Ostreidae and Sphaeriidae (Fig. 2). In Ostreidae, the β1-domain seems to have evolved from a relatively recent gene duplication, resulting in a tandem array of the two homologous β1-domains. On the contrary, the newly proposed P. coreanum αβ1β1-MT shows no apparent sequence homology between the two β1-domains. Possibly, the multiplication of β1-domain in Sphaeriidae might have been an earlier divergent event. Currently, the evolutionary route for the acquisition of additional β1-domain in P. coreanum MT is open to hypothesize. One possible scenario is the duplication of β1-domain from the prototypic αβ1-MT, followed by further divergence in non-Cys residues. Alternatively, the other possibility is that the more ancestral β1β1-MT (not yet reported in extant bivalve MTs) might have acquired additional α-domain through the duplication to β1β1β1-MT followed by the conversion of one of β1-domains to α-domain (Fig. 2). Molecular phylogeny of bivalve α-domains also shows that P. coreanum α-domain is independently placed without being affiliated with any other bivalve α-domain ortholog in a subclade (Additional file 1: Figure S8). Hence, further efforts to exploit paralogue isoforms from this species (P. coreanum) and/or similarly structured orthologs from its closely related species should be needed to get a deeper insight into the mechanism responsible for the emergence of αβ1β1-MT in Sphaeriidae.
From the same molecular phylogenic tree, putative domains from the two A. irradians MT isoforms (Liu et al. 2006; Wang et al. 2009) are positioned in either the main β1-clade or a small clade consisting of only A. irradians MT domains (Fig. 3). Two C-terminal domains respectively from A. irradians MT1 and MT2 are placed in the main β1-clade. On the other hand, the small clade are exclusively comprised of three putative β2-like domains from the MT1 (first to third domains predicted from the N-terminal side) and two from MT2 (first and second domains). A. irradians MTs represent non-canonical shape that does not perfectly match the known typical β-domain structure. Nevertheless, if the Cys distribution pattern is fundamentally considered, the multiplied domains in A. irradians MTs could be classified as β2-like structure (possibly designated β2′-domain). Hence, the overall domain structures of A. irradians MT1 and MT2 could be designated β2′β2′β2′β1-MT and β2′β2′β1-MT, respectively. In bivalve class, the MT proteins possessing multiple β-domains without α-domain have been reported only in the Crassostrea β2β2-MT-IIIs, and this structure has been considered to have developed from the early duplication of the ancestral β2-MT. However, A. irradians MTs represent a C-terminal β1-domain as well as the multiple β2′-domains (Fig. 2). Two plausible, but untested, hypotheses may be possible regarding the evolution of such unusual domain structure in A. irradians MTs. One is the acquisition of C-terminal β1-domain in the ancestrally duplicated β2β2-MT (i.e., giving rise to β2β2β1-MT) followed by further divergent process resulting in current shapes of A. irradians MT isoforms. The other, more plausible, hypothetical path is the multiplication of β2-domain from the prototypic β2β1-MT structure that is seen in most gastropod MTs (Jenny et al. 2016; Palacios et al. 2011). As described above, the present study has already noted that the multiplication of β2-domain (i.e., β2nβ1-structure) might have been one of divergence mechanisms in certain gastropod taxa (Fig. 2). However, unlike the tandem duplication of homologous β2-domain in gastropod MTs, A. irradians MTs represent little sequence similarity among β2′-domains, suggesting that the multiplication of these β2′-domains in each A. irradians MT isoform may not be the recent duplication event. Although need to be further challenged, the ancestral β2β1-MT in the phylum Mollusca perhaps might have gone through two separate paths (Fig. 2). Namely, one might have been a preservative path from the ancestral β2β1-MT to produce the conserved β2β1-structured MTs which seen in most extant gastropod MTs, although some exceptional gastropod species represent additional lineage-specific, recent duplication(s) of homologous β2-domain. On the other hand, the other path might involve earlier duplication of β2-domain from the ancestral β2β1-MT, giving rise to multiple β2-like domains linked to the C-terminal β1-domain in certain bivalve taxa. Based on this, the extant shapes of A. irradians MT paralogs may reflect the consequences of differential rounds of β2-domain duplication from an ancestral β2β1-MT. However, further validation is needed to test whether this divergent process might have occurred before or after speciation events in the Pectinoida lineage.
What are the functional or physiological implications of domain duplications (or multiplication) in molluskan MTs? The evolution of larger MT proteins has been proposed as a strategic means that might likely be advantageous for benthic organisms that are believed to experience a greater exposure to metals due to their ecological niche (Jenny et al. 2004; Jenny et al. 2006; Tanguy and Moraga 2001; Tschuschke et al. 2002). Although the paucity or limitation of the functional studies on such large MTs hurdles to hypothesize comprehensively the relevant mechanism(s) in detail, there have been some hypothetical evidences or suggestive assumptions. First, based on the heterologous expression assay by using the recombinant microbial systems, a couple of noteworthy experiments have shown that large-sized multi-domain-structured MT proteins would be able to confer greater Cd resistance of the hosts (Tanguy et al. 2001; Tschuschke et al. 2002). Second, several independent previous studies have claimed that amplification and/or tandem duplications of MT genes might have been an advantageous process to attain the strengthened ability of metal tolerance (Cho et al. 2009; Beach and Palmiter 1981; Maroni et al. 1987; Mehra et al. 1990; Stephan et al. 1994). Because the number of Cys residues in MT proteins has been taken into account as a fundamental factor to determine the number of metal ions bound or reserved by the MTs (Amiard et al. 2006), more Cys attained by domain duplications might be beneficial in the conference of metal resistance in a broad sense. Third, domain multiplication(s) accompanied with significant substitutions/replacements of non-Cys residues could offer a chance to confer some novel functions on large MTs. Because aa replacements on non-Cys residues in MT have been known to represent significant effects on metal-binding specificity and kinetic reactivity (Palacios et al. 2011; Pedersen et al. 1994; Kurasaki et al. 1997; Munoz et al. 2000), such a divergent pattern (domain duplication together with significant aa substitutions) might also increase the kinds of metals reacted by these MT proteins. Taken together, the ancestral or prototypic MT protein has diverged into various isoforms with a great structural diversity in the phylum Mollusca. Structural diversifications driven by both domain duplication and aa replacements might have led certain subfunctionalization and/or neofunctionalization of MT proteins in an isoform-dependent fashion (Tanguy and Moraga 2001).
Conclusions
Phylum Mollusca represents a great structural diversity of MT, a core suite playing key roles in both homeostatic regulation of essential metals and detoxification of trace metals in living organisms. The structural diversity of molluskan MTs have been achieved essentially through the domain duplication events from an ancestral, singular domain-MT. Domain duplication have been followed by further diversification and selection toward needs for acquiring metal selectiveness, specialized novel function, and improved capacity of metal homeostasis/detoxification. With this viewpoint, novel paths for domain divergences of some gastropod and bivalve MT families proposed in this review could shed new light onto the revision and update of the hypothesis for evolutionary differentiation of MTs in the molluskan lineage.