Background
The genus Hippocampus includes 41 species throughout the world (Lourie et al. 2016). Hippocampus kuda (common seahorse) has an enormous distribution, including the Indo-Pacific Ocean, except for the eastern Pacific. Some populations are mature at 7 cm in standard length (SL), whereas others grow to 17 cm SL. Their color varies: yellow, sandy, or white, but usually black, with a grainy texture or dark spots. The snout is thick, and the coronet is overhanging at the back and often topped by a slight depression (cup-like). A single eye spine is prominent, but body spines are low and blunt (rounded bump only). H. kuda is a mainstay species in the aquarium trade and traditional Chinese medicine. In contrast, H. reidi (slender seahorse) is distributed along the western Atlantic coast, from the USA to Argentina. Adults measure from 10 to 18 cm SL. Their color also varies: red, yellow, orange, black, or brown, with white saddles and scattered black dots. The snout is clumpy, and the coronet is convoluted (like a crumpled ball of paper), rounded, and often folded in upon itself. Double eye spines are prominent, but body spines are none or low, and blunt . H. reidi is used in the aquarium trade and as curios (dried specimens) (Lourie et al. 1999; Indiviglio 2002; Lourie et al. 2004; Hercos and Giarrizzo 2007; Piacentino 2008; Lourie 2016; Lourie et al. 2016).
Although their reproductive rates are low and their home ranges limited, H. kuda and H. reidi are generally valuable seahorse species, with worldwide market demand, which may cause their overexploitations and threaten natural populations through the undoubted pressure of the activities of fisheries that supply the market (Vincent 1996; Lourie et al. 1999). From a conservation perspective, aquaculture is an important way to conserve endangered species and bioresources from extinction (Ireland et al. 2002; Lindberg et al. 2013; Ho et al. 2015). However, interspecies hybridization can be a management problem to design plans such as conservation of biodiversity and process of breeding (Allendorf et al. 2001; do Prado et al. 2011; Morgan et al. 2012; Brennan et al. 2015; Ho et al. 2015). Therefore, interspecific hybridization study is necessary in the context of aquaculture to ensure effective bioresource management. With rare records of interspecific hybridization in the family Syngnathidae, according to a review by Ho et al. (2015), four cases of interspecific hybridization in the seahorse have been reported: H. kuda chesteri (suspected ♂ H. reidi × ♀ H. kuda) (Bull and Mitchell 2002; Woodall et al. 2009), ♂ H. algiricus × ♀ H. hippocampus (Otero-Ferrer et al. 2015), ♂ H. erectus × ♀ H. reidi, and ♀ H. erectus × ♂ H. reidi (Ho et al. 2015). A large seahorse database of the mitochondrial DNA (mtDNA) cytochrome b sequences is available for intra- or interspecies comparisons in morphological or molecular ways from its phylogenetic investigation (Casey et al. 2004; Lourie et al. 2005; Han et al. 2017a, b; Woodall et al. 2018). Nuclear DNA (ncDNA) ribosomal protein S7 gene is also useful for species-level identification; however, the S7 gene of H. reidi revealed gene introgression between H. reidi and H. erectus, although the two species are more genetically distant than H. kuda and H. reidi (Teske et al. 2004; Ho et al. 2015).
In the present study, we induced hybridization of artificially reared ♂ H. kuda and ♀ H. reidi, and describe the differences between the ♂ H. kuda × ♀ H. reidi hybrid and the parental species, and our new morphological description with molecular verification will help for the better understanding of taxonomic information for future conservation effort.
Methods
Juveniles were raised in separate rectangular glass tanks (50 × 30 × 28 cm), connected to filter tanks (26 × 26 × 28 cm). The tanks were monitored daily, and any uneaten food was siphoned away as waste. We cycled the tank water 9–10 times per day. The rearing conditions were as follows: temperature 24.0 ± 0.5 °C, salinity 33 ± 1.0 ppt, pH 7.78 ± 0.42, dissolved oxygen 6.8 ± 0.3 mg/L, and photoperiod 14 h light:10 h dark. Lebistes reticulatus (1–2 days old), 5–7 mm copepod nauplii of 2–3 weeks (O.S.I., Snowville, Utah, USA), and blood worms were fed daily to the juvenile seahorses (Choi et al. 2006).
We housed together only male H. kuda and female H. reidi purchased from markets in Korea in the spring of 2014. Under daily monitoring, we sampled 0- to 45-day old hybrid (♂ H. kuda × ♀ H. reidi) offspring (n = 133) and 0- to 45-day old juveniles of H. kuda (n = 77) to compare the early growth patterns of them. The meristic counts of seven broodstock of H. kuda (n = 5) and H. reidi (n = 2) were compared with the hybrid and H. kuda offspring for morphological analysis. Six specimens were used for a molecular analysis: one each of the H. kuda and H. reidi broodstock, two H. kuda offspring, and two hybrid offspring. We preserved and fixed the specimens in 99% ethanol or 10% formaldehyde.
We counted the meristic characters, including the trunk rings, tail rings, dorsal fin rays, pectoral fin rays, anal fin rays, eye spines (supraorbital spine), cheek spines (infraoperculum spine), and nose spines (anteorbital spine). However, we could not check the tail rings of one of the H. reidi broodstock specimen, because it had a damaged tail tip. Spine counts have limited utility in the very early stage of growth because they only mature after a certain stage. Whereas, a melanophore pattern analysis is an alternative method in this early life stage and has been used as a classificatory key in the early life stages of many other fish species, although the pattern disappears retrogradely as growth proceeds (Matarese et al. 1989).
Measurements were made with the microscope-integrated Active Measure software (Shinhanoptics, Seoul, Korea) based on set points for the following parameters: head length (HL), trunk length (TrL), tail length (TaL), snout length (SnL), snout depth (SnD), head depth (HD), dorsal and pectoral fin base lengths (DfL and PfL, respectively), eye diameter (ED), and standard length (SL) (Lourie 2003; Choo and Liew 2006). We derived age–SL relational expression and allometric relational expression (SL–HL, SL–TrL, SL–TaL, SL–SnL, SL–SnD, SL–HD, SL–DfL, SL–PfL, and SL–ED) using LOESS (local regression) curves in ggplot2 package of R software ver. 3.3.1 (Jacoby 2000; Wickham 2009; R Core Team 2017), and we examined the differences in the growth patterns of the seahorses by LOESS, a non-parametric approach of simple polynomial regression represented by a moving average. Because the value of the regression function for the point is obtained from local polynomial, LOESS do not give simple mathematical formula and not predict movements before and after the regression of the data. Nonetheless, LOESS visualize flexible fitting regression, and it is more understandable for moving trend in the sample than many other methods. We set the 98% confidence intervals of LOESS curve on the means, and a two-tailed P value is considered as statistical significance of rejection for the regression (Jacoby 2000; Lim et al. 2013).
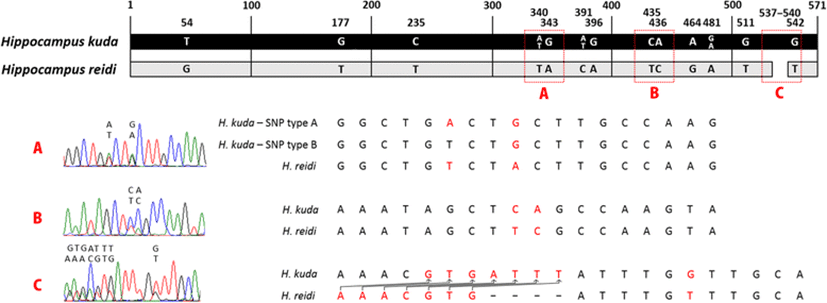
Genomic DNA was extracted from the right eyeball or right-side tail tissue in all four specimens using the AccuPrep® Genomic DNA Extraction Kit (Bioneer, Daejeon, Korea). We used the mtDNA cytochrome b and ncDNA ribosomal protein S7 gene sequences for the genetic analysis. We compared the sequences with the GenBank sequences (http://www.ncbi.nlm.nih.gov/) for subclades A and C of H. kuda (Lourie et al. 2005) and H. reidi (Teske et al. 2007) to identify the species, using H. trimaculatus as the outgroup (Chang et al. 2013).
PCR was performed on an S1000™ Thermal Cycler (Bio-Rad, Hercules, California, USA) in a reaction mixture containing 3 μl of 10× Ex Taq buffer (plus 20 mM Mg2+), 2.4 μl of 2.5 mM dNTPs, 1 μl of forward primer, 1 μl of reverse primer, 0.1 μl of TaKaRa Ex Taq DNA polymerase (Takara Bio, Kusatsu, Shiga, Japan), 3 μl of genomic DNA, and distilled water to a total volume to 30 μl. The PCR was designed to amplify the mtDNA cytochrome b gene using primers Shf2 (5′-TTGCAACCGCATTTTCTTCAG-3′) and Shr2 (5′-CGGAAGGTGAGTCCTCGTTG-3′) under the following conditions: initial denaturation at 94 °C for 2 min 30 s, 35 cycles of denaturation at 94 °C for 30 s, annealing at 50 °C for 30 s, and extension at 72 °C for 75 s, with a final extension at 72 °C for 5 min (Lourie and Vincent 2004), and ncDNA ribosomal protein S7 gene (1st intron, RP1) using primers S7RPEX1F (5′-TGGCCTCTTCCTTGGCCGTC-3′) and S7RPEX2R (5′-AACTCGTCTGGCTTTTCGCC-3′) under the following conditions: initial denaturation at 95 °C for 1 min, 30 cycles of denaturation at 95 °C for 30 s, annealing at 60 °C for 1 min, and extension at 72 °C for 2 min, and a final extension at 72 °C for 10 min (Chow and Hazama 1998). The samples were purified with a LaboPass™ PCR Purification Kit (Cosmogenetech, Seoul, Korea). The sequencing reactions were performed in a DNA Engine Tetrad 2 Peltier Thermal Cycler (Bio-Rad) using an ABI BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Waltham, MA, USA). The sequences were aligned with BioEdit version 7 (Hall 1999). Genetic distances were calculated with the Kimura two-parameter model (Kimura 1980) in the MEGA version 6.05 software (Tamura et al. 2013). A neighbor-joining tree was constructed from 696 bp of the cytochrome b gene using MEGA, and confidence levels were assessed with 1000 bootstrap replications. Heterozygosity of ncDNA causes a mixed signal as double peaks of sequence chromatograms, and single nucleotide polymorphism (SNP) and insert/deletion (indel) overlap may reveal hybridization (Sousa-Santos et al. 2005; Sonnenberg et al. 2007; Bae et al. 2016). Therefore, 571 bp of ribosomal protein S7 gene via forward and reverse reading was used for demonstration of hybridization in this study (Fig. 1).
Results
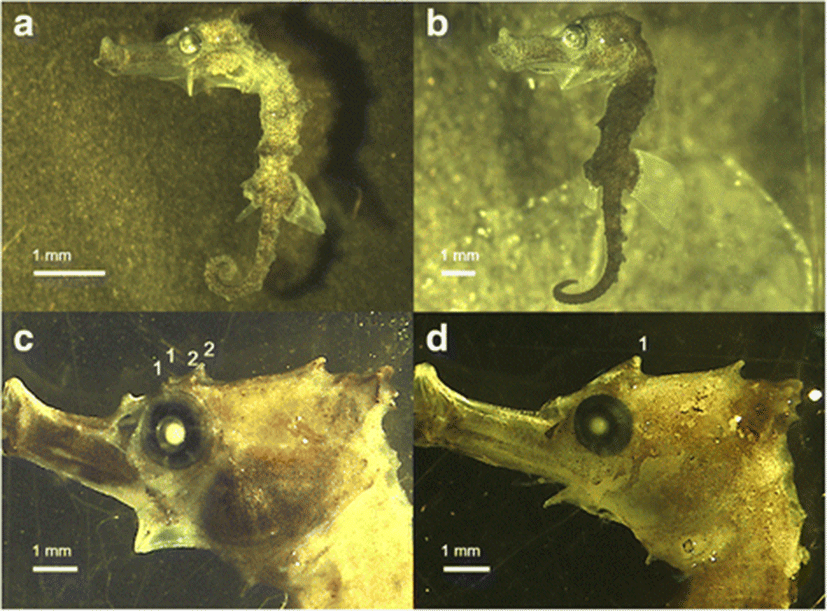
We observed no significant morphological differences in the trunk rings, tail rings, dorsal fin rays, pectoral fin rays, anal fin rays, nose spines, and cheek spines between H. kuda and the hybrid, because the ranges of these features overlapped (Table 1). However, all 1- to 10-day old specimens of the hybrid had dense melanophores only on the tails, and their striped trunk rings showed an intermediate form (Fig. 2a) compared with those of the parents. H. kuda has dense melanophores on its whole body or only on the trunk and tail, whereas the juveniles of H. reidi have thin melanophores or a striped pattern and the melanophores only occur on their prominent trunk and tail rings (Fig. 2b; see Choo and Liew 2006; Mai and Loebmann 2009; Van Wassenbergh et al. 2009). However, the melanophores of the hybrids became more like the dense melanophores on the whole bodies of H. kuda after 11 days, so the new pattern was limited to the early stage of development. In contrast, the two eye spines (or their traces) in the hybrid were apparent after they were 9 days old, which were not inherited from H. kuda (one eye spine), but from H. reidi (two eye spines) (Table 1).
|
Present study |
Lourie et al. (2004) |
|||||
|---|---|---|---|---|---|---|
|
Juvenile (offspring) |
Adult (broodstock) |
Adult |
||||
|
Hybrid (♂ H. kuda × ♀ H. reidi) |
H. kuda |
H. reidi |
H. kuda |
H. reidi |
H. kuda |
|
|
Number |
133 |
77 |
2 |
5 |
39** |
80** |
|
Trunk ring |
11 |
11 |
11 |
11 |
11 |
11 |
|
Tail ring |
34–38 (36) |
35–37 (36) |
32 |
35–37 |
31–39 (35) |
34–38 (36) |
|
Dorsal fin ray |
15–20 (17) |
16–19 (17) |
16–17 |
16–18 |
16–19 (17) |
17–18 |
|
Pectoral fin ray |
15–18 (16) |
15–17 (16) |
15–16 |
15–17 |
15–17 (16) |
15–18 (16) |
|
Anal fin ray |
4 |
4 |
4 |
4 |
– |
– |
|
Eye spine* |
2 |
1 |
2 |
1 |
1–2 (2) |
0–1 |
|
Nose spine* |
0 |
0 |
0 |
0 |
– |
0 |
|
Cheek spine* |
1 |
1 |
1 |
1 |
1–2 (2) |
1–2 |
*Spine values are represented by the day after fixation. Bracket indicates mode in the count range
**Number of specimens is stated in Lourie et al. (1999)
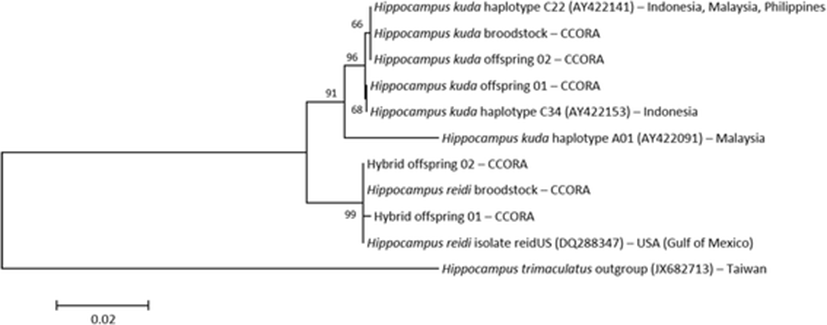
One of the H. reidi sequences from Teske et al. (2007) was most similar with the H. reidi sequences used in our study (genetic distance: 0.000–0.001), and haplotypes C22 and C34 of H. kuda in Lourie et al. (2005) was identical to our H. kuda sequences (Fig. 3). The distance between subclades A and C of H. kuda was 0.025–0.026, and the distance between the subclade C and H. reidi was 0.025–0.028, and the distance between the subclade A and H. reidi was 0.042–0.043. The outgroup distances were 0.174–0.176 with the subclade C, 0.189 with the subclade A, and 0.174–0.175 with H. reidi, respectively. Thus, the maternal molecular mtDNA data indicated that the hybrid offspring corresponded to H. reidi and that the eye spine phenotype was inherited from H. reidi, although the hybrid specimens were born from the male H. kuda brood pouch (Fig. 3). Analysis of S7 sequences with a length of 571 bp revealed an average of one indel and 13 SNP overlaps, and these overlaps representing double peaks in the sequence also demonstrated the hybridization of both species (Fig. 1).
We confirmed the growth rates of both species increased most rapidly between 20 and 25 days; therefore, we distinguished two phases of development in both species (Fig. 4). According to the significant differences (P < 0.02) of the SL–age relationship, the first phase was identified between 3 and 18 and the second between 24 and 45 days old. The slopes of both species from 3 to 18 days old are almost parallel, but between 24 and 45 days old, species exhibit different growth rates (Fig. 4a). Therefore, the growth rates of the hybrid and H. kuda were different in both phases. Our results supported the multilinear graph of the H. kuda growth rate reported by Choo and Liew (2006). In this study, the linear regression equation from first section of hybrid was y = 0.3495x + 8.0311 (coefficient of determination [r2]: 0.8235), and it from first section of H. kuda was y = 0.3147x + 6.7682 (r2: 0.7865). After that, it from second section of hybrid was y = 1.2677x − 13.143 (r2: 0.9252), and it from first section of H.kuda was y = 1.0336x − 10.059 (r2: 0.9207). The allometric alteration was separated by 15 mm SL standard in the present study as a result of growth turning point versus the middle of 20 mm SL of Choo and Liew (2006), so the patterns were not completely same (Fig. 4; Choo and Liew 2006). Nonetheless, our result supported the previous study that the second phase grew faster than the first phase in H. kuda when they entered the explosive second phase of growth.
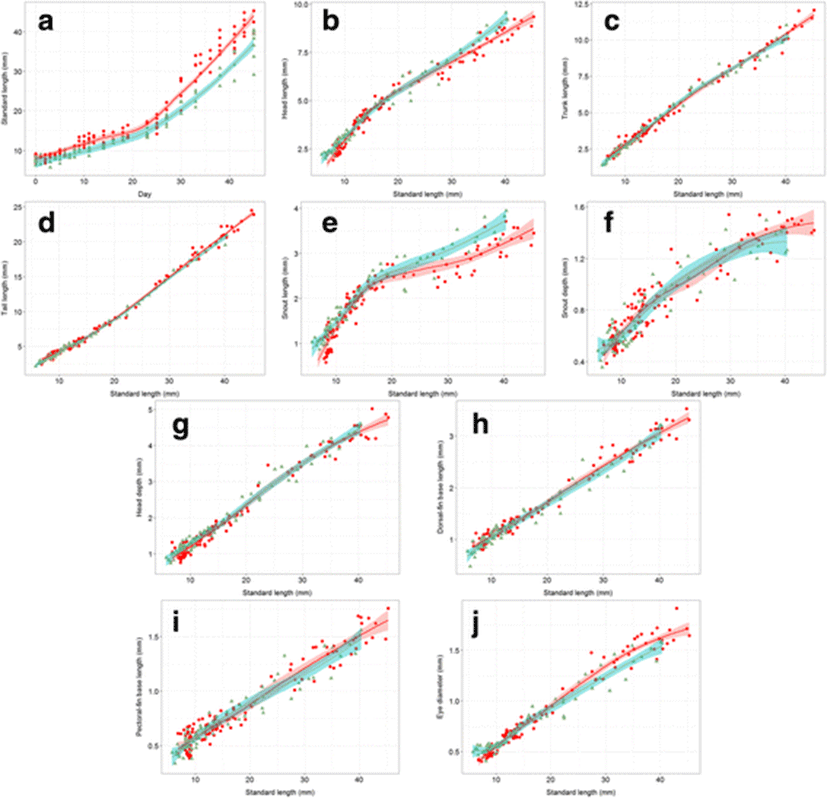
Most of the allometric graphs for the hybrid and H. kuda showed non-significant differences, except for HL (before 9 mm SL and after 33 mm SL), SnL (before 10 mm SL and after 28 mm SL), and ED (before 8 mm SL and after 27 mm SL). The measurements that differed according to LOESS (P < 0.02) were some related to the head, indicating that these features have different growth patterns in the hybrid and H. kuda (Fig. 4b–j). In contrast, measurements of TrL, TaL, SnD, HD, DfL, and PfL did not differ on LOESS analysis, suggesting that it is difficult to distinguish them based on the allometric patterns in these traits. In two of the three measurements that did change after hybridization (HL, SnL), the curves for allometric growth were higher level for H. kuda than for the hybrid. However, the slope for one of these three measurements (ED) was steeper in the hybrid than in H. kuda (Fig. 4b–j).
Discussion
The examined two species, H. kuda and H. reidi, are known to have different morphotypes in previous studies (Lourie et al. 1999; Lourie et al. 2004; Lourie 2016). However, the morphological characters of some seahorses can be ambiguous because wide meristic or morphometric ranges occur in these characters within the same species, and their ranges can overlap among different species, including H. kuda and H. reidi (Hubbs 1922; Lourie et al. 1999, 2004; Ho et al. 2015). In this study, eye spine and melanophore are useful tools to distinguish the two seahorses in their early stages, and thus, these tools will help to quickly identify the hybrids. Furthermore, genetic tools are also very useful to improve identifying species and intraspecific hybrids as improved taxonomic analysis (do Prado et al. 2011; Ho et al. 2015). Although mtDNA is a matrilineal inheritance system, the discordance between an intermediate or patrilineal phenotype and the molecular results paradoxically confirms interspecific hybridization (Wayne and Jenks 1991; Kwun and Kim 2010). Moreover, the hybrid sequence showed double peaks on SNP site before the overlapping indel site which continued throughout the sequence. Therefore, this heterozygosity demonstrates the hybridization of both species (Fig. 1).
A shorter snout allows the seahorse to successfully capture concentrated prey and to use its pivot-feeding strategy to catch evasive prey (Leysen et al. 2011; Van Wassenbergh et al. 2011), and a bigger eye closely related to favorable vision of feeding except for fishes having smaller eyes in dark or murky environments (Gatz 1979; Caves et al. 2017). These points support that hybridization between the two species improved the growth rate by altering the snout length and eye diameter. This improvement may be genetically inherited from H. reidi; however, this must be confirmed with overall comparison of the hybrid and H. reidi offspring to determine whether the phenomenon is influenced by intermediate type or synergy.
Molecular evidence of monogamy has been reported in the many seahorse species in both laboratory and wild populations, including H. kuda and H. reidi (Rosa et al. 2007; Freret-Meurer and Andreata 2008; Ooi et al. 2010; Rose et al. 2014). However in this experiment, polyandry occurred among one H. reidi (♀) and several H. kuda (♂) specimens before interspecific fertilization, even though the seahorses are known to be monogamous species. Polygamy has already been reported in several seahorse species in nature (Kvarnemo et al. 2000; Foster and Vincent 2004); and thus, we newly report that polygamy between the two species can also occur in laboratory conditions.
The genus Hippocampus has been listed in Appendix II of the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES 2017), and international trade is restricted (Vincent et al. 2011). In Asia, even if trade is approved, H. reidi must be transported from its place of origin to a lucrative market over the great distance with exposure of high mortality; therefore, the commercial distribution of H. reidi is limited. Nevertheless, the conservation plans may be revised or extended for H. kuda and H. reidi, because the definition of the name H. kuda has been controversial with its sister species (Lourie et al. 1999; Teske et al. 2005; Lourie et al. 2016) and both species have a possibility of interspecific hybridization in distribution channels for economic benefits. The distinct morphotypes, geographic isolation, and genetic results can confirm that H. kuda and H. reidi are separate species, or they may be the products of the ongoing evolutionary divergence of a single complex (Teske et al. 2005; Lourie et al. 2016).
Chester Zoo (UK, http://www.chesterzoo.org/) breeds H. reidi and H. kuda, and its researchers insist that H. kuda chesteri can reproduce (Bull and Mitchell 2002; Woodall et al. 2009). However, hybrid verification of the H. kuda chesteri is not fully conducted because mtDNA verification shows the maternal result (Woodall et al. 2009). Therefore, we must check the capacity of H. kuda chesteri because it increases the importance of the hybrid between female H. kuda and male H. reidi produced with the opposite mating strategy in the present study. Interactive sexual hybridization also doubts that the species are capable of full genetic exchange.
Conclusions
Discordance between morphological results (melanophore and development patterns) and the molecular result of mtDNA cytochrome b (neighbor-joining tree) paradoxically confirmed interspecific hybridization of two seahorses, H. kuda and H. reidi. Moreover, heterozygosity of ncDNA ribosomal protein S7 gene via partially mixed template also supported the hybridization. In allometric growth comparison, the snout length growth was slower, but the eye diameter growth was faster in hybrids than those of H. kuda, which suggests successful suction has a favorable impact in the early stage growth. A detailed morphological study is essential for the immediate analysis of these species and to support their future management. Improved taxonomic information will aid in distinguishing hybridization from parental phenotypes so as to monitor hybrids in the international trades.