Background
Olive flounder is one of the most important aquaculture fish in the Republic of Korea. The production of olive flounder in aquaculture has been greatly threatened by the increase in contamination of the environmental with various microbial pathogens, including bacteria, virus, and parasites (Kim et al. 2010). For sustainable development of aquaculture industries producing olive flounder, proper strategies for management of diseases affecting this fish species are desired. As of date, several studies have focused on the prevention of pathogenic diseases in olive flounder. However, studies related to the management of viral diseases have been relatively scarce.
Viral hemorrhagic septicemia virus (VHSV) belong to the genus Novirhabdovirus, family Rhabdoviridae, and cause severe damages in various farmed fishes including olive flounder, salmon, rainbow trout, turbot, and freshwater species (Mortensen et al. 1999; Schutze et al. 1999). VHSV is a bullet-shaped single-stranded RNA, which encodes six proteins consisted in a nucleoprotein (N), a phosphoprotein (P), a matrix protein (M), a glycoprotein (G), a non-virion protein (NV), and a polymerase (L), and the genome is approximately 11,000 nucleotides long (Einer-Jensen et al. 2004). Based on a phylogenetic analysis of the N, G, and NV genes sequence, VHSV can be grouped into four major genotypes (genotype I: European; genotype II: Baltic sea; genotype III: North Atlantic Sea; genotype IV: North American and Korean/Japanese) which showed geographical distribution (Einer-Jensen et al. 2004; Lumsden et al. 2007).
Stimulator of interferon gene (STING), also known as mediator of interferon regulatory factor 3 (IRF3) activation (MITA) (Zhong et al. 2008), plays an essential role in the host immune defense mechanisms, particularly against viral infections, by expediting the innate immune signaling. Various studies have reported the effect of STING on viral infections (Nakhaei et al. 2010; Aguirre et al. 2012). STING-knockout mice were found to be highly vulnerable to infection with vesicular stomatitis virus (VSV) (Ishikawa et al. 2009). In addition, STING-mediated antibacterial response was also reported in mammals (Jin et al. 2013). STING is a transmembrane protein localized in the endoplasmic reticulum (ER) of various types of cells, including the antigen-presenting cells, such as macrophages and dendritic cells, as well as in the endothelial and epithelial cells (Ishikawa and Barber 2008; Barber 2011). The overexpression of STING triggers the activation of both nuclear factor kappa B (NF-kB) and interferon regulatory factor 3 (IRF3) and thereby induces the production of type I interferon that triggers the host immune response (Zhong et al. 2008; Ishikawa et al. 2009; Abe and Barber 2014). Further, STING is involved in the phosphorylation of signal transducer and activator of transcription 6 (STAT6) via TANK-biding kinase 1 (TBK1) without association of janus kinases (JAKs) (Chen et al. 2011). In addition, STING functions as a pattern recognition receptor (PRR) for some cyclic dinucleotides, such as cyclic diguanylate monophosphate (c-di-GMP) (Burdette et al. 2011).
STING orthologs from several fish species were identified and characterized to show their functional aspects (Sun et al. 2011; Feng et al. 2014; Ge et al. 2015; Huang et al. 2015). However, few studies have been reporting the role of STING orthologs from marine fish species. In the present study, we cloned and structurally characterized a STING ortholog (PoSTING) from the olive flounder, Paralichthys olivaceus. We also analyzed the transcriptional expression of STING and type I interferon upon artificial infection of the olive flounder with virus and bacteria.
Methods
To identify the cDNA sequence of PoSTING, degenerate primers were designed from within the highly conserved nucleotide regions of STING sequences from Stegastes partitus (XM_008282192.1), Haplochromis burtoni (XM_005916606.1), Maylandia zebra (XM_004563199.1), and Xiphophorus maculatus (XM_005811123.1). Polymerase chain reaction (PCR) was performed using the designed degenerate primers (forward: 5′-AAGAAGAACGTAGCCCACGG-3′, reverse: 5′-AGAACTCCTCTCTCTCCTGC-3′), and the partial sequence was cloned. The acquired partial sequence was used for the designing of gene-specific primers for rapid amplification of cDNA ends (RACE). To acquire the full-length cDNA sequence of PoSTING, RACE was performed using CapFishing™ Full-length cDNA Premix kit (Seegene, South Korea), according to the manufacturer’s instructions. The PCR products were visualized on a 1% agarose gel and purified using the GEL & PCR Purification system (BIOFACT, South Korea). Subsequently, the purified PCR product was ligated into T-Blunt vector, according to the protocol provided with the T-Blunt™ PCR cloning kit (SolGent, South Korea), and the generated construct was transformed into Escherichia coli DH5α competent cells. Finally, the plasmid with correct insertion was purified using the SolGent Plasmid Mini-Prep kit (SolGent, South Korea) and sequenced.
The full-length cDNA sequence of PoSTING was analyzed using the Basic Local Alignment Search Tool (BLAST) available through the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/blast). The open reading frame (ORF) was determined by UGENE software. The deduced amino acid sequence and the physicochemical properties of the predicted protein were identified using the UGENE software. The anticipated domain architecture was predicted by Simple Modular Architecture Research Tool (SMART) (http://smart.embl-heidelberg.de/). Phylogenetic tree was constructed based on the deduced amino acid sequence of PoSTING and STING orthologs from other species, using the neighbor-joining (NJ) algorithm embedded in MEGA 5.3 program (Tamura et al. 2011). Furthermore, a 3D homology model of PoSTING was predicted by SWISS-MODEL server (https://swissmodel.expasy.org/) and visualized using PyMOL software.
Healthy olive flounder fish (with an average body weight of 50 ± 6 g) were maintained in 150-L tanks with filtered seawater and continuous aeration at a temperature of 18 ± 1 °C, then used to investigate the tissue distribution of STING mRNA. The temperature of seawater in the tank for viral hemorrhagic septicemia virus (VHSV)-challenged group was maintained at 14 ± 1 °C, then used to analyze the STING and IFN-I expression upon VHSV challenge. All the fish were acclimatized for 1 week prior to the experiments. To investigate the tissue distribution of PoSTING transcripts, 14 different tissues including spleen, head kidney, kidney, gonad, muscle, gill, blood, skin, brain, eye, heart, intestine, stomach, and liver were collected from three fish. To harvest the blood cells, the blood was collected and immediately centrifuged at 3000×g for 10 min at 4 °C. All the tissues isolated were snap frozen in liquid nitrogen and stored at − 80 °C until use.
For the immune challenge experiment, Edwardsiella tarda and VHSV were injected intraperitoneally in the fish. E. tarda, stored at − 80 °C as glycerol stocks, were plated onto a brain heart infusion (BHI) agar plate and incubated at 25 °C for 25 h. A single colony was incubated in 5 mL of BHI broth with agitation at 25 °C for 4 h. The cultured bacteria were centrifuged at 2000×g for 20 min, and the pellet obtained was washed using 1X phosphate-buffered saline (PBS). The final concentration of bacteria was adjusted to 104 CFU/100 μL/fish. For the viral challenge experiment, VHSV was grown in the fathead minnow (FHM) cell line with the Minimum Essential Medium Eagle (Sigma, USA). Virus were harvested and resuspended at a concentration of 1 × 108 median tissue culture infectious dose (TCID50)/100 μL/fish. Hundred microliters of E. tarda and VHSV were injected intraperitoneally in different groups of fish (n = 30/group). An equal volume (100 μL) of PBS was administered to fish in a different group that was used as a control. Four fish from each group were randomly selected and dissected at different time intervals of 0-, 5-, 10-, 24-, 48-, and 72-h post injection to isolate the kidney tissues. All the isolated tissues were snap frozen in liquid nitrogen and stored at − 80 °C until RNA extraction.
Total RNA was extracted from the isolated tissues (see the “Experimental animals and tissue collection” and “Challenge experiment” sections) using RNAiso Plus (TaKaRa Bio Inc., Japan), according to the manufacturer’s protocol. The concentrations and purities of the extracted RNA samples were assessed using a spectrophotometer (NanoDrop 2000C, Thermo Scientific, USA) by measuring the absorbance at 260 and 280 nm. The A260/280 ratio of the extracted RNA samples was over 1.8. Furthermore, the integrity of the RNA samples was confirmed by agarose gel electrophoresis. To prevent genomic DNA contamination, DNase treatment was done using an RQ1 RNase-Free DNase kit (Promega, USA), as per the manufacturer’s instructions. All the RNA samples were kept at − 80 °C until use.
The quantitative real-time PCR (qPCR) analysis was performed on Thermal Cycler Dice™ Real-time System TP850 (TaKaRa Bio Inc., Japan) to quantify the level of mRNA expression of PoSTING. The gene-specific primers used to amplify the PoSTING fragment were 5′-CTTGGGGTCACGGCTCCAAGAAG-3′ (forward) and 5′-GCCGAGTCTACAAGCACAGCGT-3′ (reverse) and those used to amplify the internal reference gene (accession no. AB915949.1), olive flounder elongation factor 1 alpha (PoEF1α), were 5′-GCAGCTCATTGTTGGAGTCA-3′ (forward) and 5′-ACACTTGCAGGGTTGTAGCC-3′ (reverse). All the qPCRs were carried out in triplicates in a 20 μL reaction mixture containing 20 ng of total RNA, 10 μL of TOPreal™ qPCR 2X PreMIX of One-step RT qPCR kit (SYBR Green) (Enzynomics, South Korea), 1 μL of each primer (10 pmol/μL), and 7 μL of PCR grade water. The real-time PCR cycling protocol was as follows: one cycle of 50 °C for 30 min for cDNA synthesis, amplification for 45 cycles of 95 °C for 10 min, 95 °C for 5 s, 60 °C for 30 s, and 60 to 95 °C for the melting curve analysis. The baseline was set automatically by the Thermal Cycler Dice™ Real Time system TP850 program. In addition, the expression level of type I interferon transcripts was examined using gene-specific primers (forward: 5′-GAAGTGGAGGAGACTGTGGC-3′, reverse: 5′-GTGACTCACAATACAGGAGCGA-3′). The relative mRNA expression levels of the genes were analyzed by the 2-ΔΔCt method. All the data were represented as means ± standard deviation (SD), and the quantities of mRNAs were expressed relative to those of the flounder EF1α (PoEF1α) mRNA. All the PCR experiments were conducted in triplicate. Significant differences between the challenged and control groups were analyzed by GraphPad statistical software, and the P value was set as < 0.05.
Results
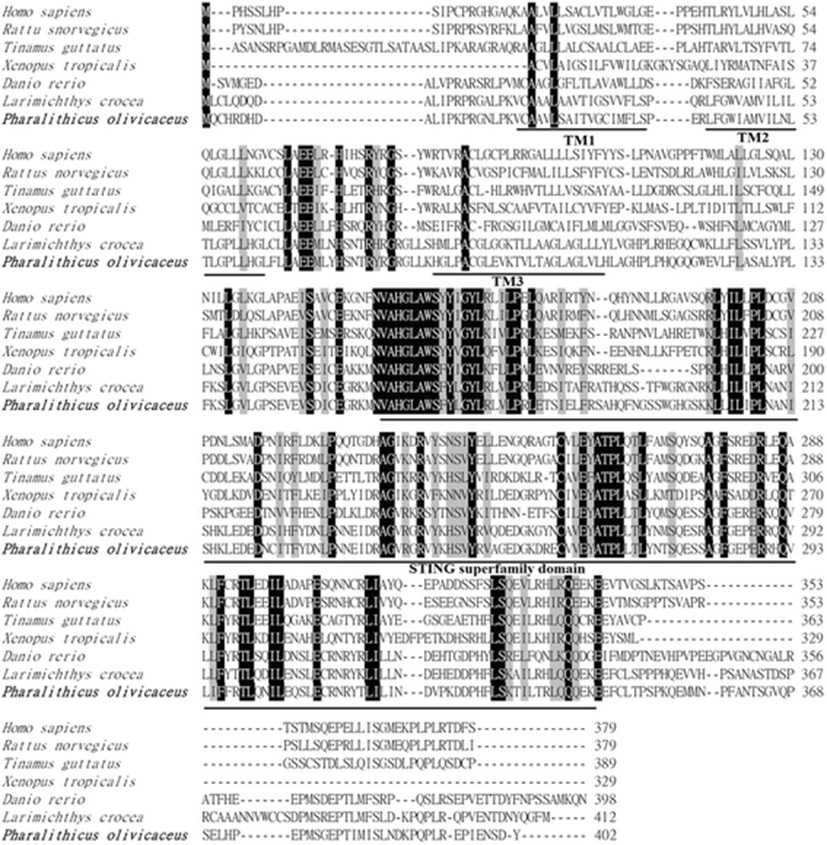
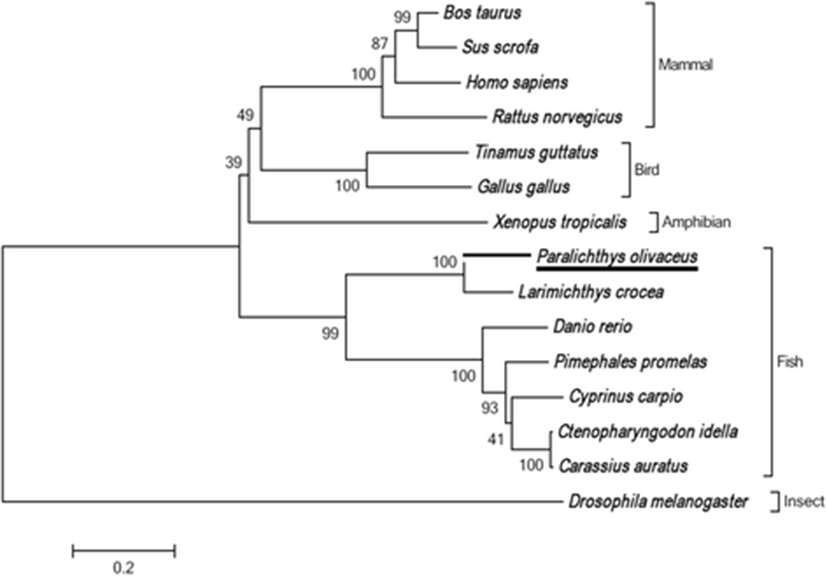
The full-length PoSTING cDNA sequence (GenBank accession number: LC148052.1) contains 1442 bp, comprising an open reading frame (ORF) of 1209 bp, a 5′-untranslated region (UTR) of 58 bp, and a 3′-UTR of 175 bp. The cDNA encoded a polypeptide of 402 amino acids, and a calculated molecular mass of 45.09 kDa was acquired using web-based software, I-TASSER (https://zhanglab.ccmb.med.umich.edu/I-TASSER/). According to in silico analysis, three possible transmembrane domains (Val21-Ser38, Leu42-Leu64, and His85-Leu107), and the characteristic STING superfamily domain (Val158-Glu342) were identified (Figs. 1 and 2). However, no signal sequence was detected at the N-terminus of PoSTING. To analyze the homology, the amino acid sequence of PoSTING was compared with those of its counterparts from other species (Table 1). The results revealed that PoSTING showed the highest identity (82.4%) and similarity (73.4%) with the Larimichthys crocea STING ortholog. In addition, PoSTING shared over 41% identity with the sequences from the other species analyzed. Multiple sequence alignment revealed comparatively higher conservation in the STING superfamily domain region, indicating functional conservation among the species (Fig. 2). The phylogenetic analysis showed two different clusters mainly separating the piscine and other higher vertebrates (Fig. 3). Olive flounder was closely clustered with Larimichthys crocea as expected, whereas the other fish species were present in a separate clade.

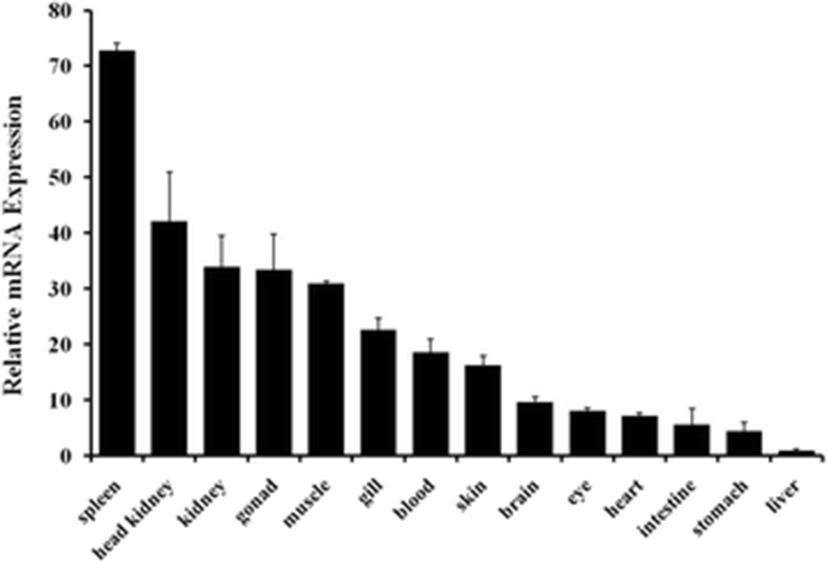
The expression of PoSTING mRNA in various tissues of healthy flounder was determined by real-time quantitative PCR. The transcripts of PoSTING were ubiquitously expressed in all the 14 tissues, with the highest expression observed in the spleen, which was over 70-fold higher than in the liver; this was followed by the expression levels in head kidney and kidney tissues. The lowest expression was observed in the liver tissue (Fig. 4).
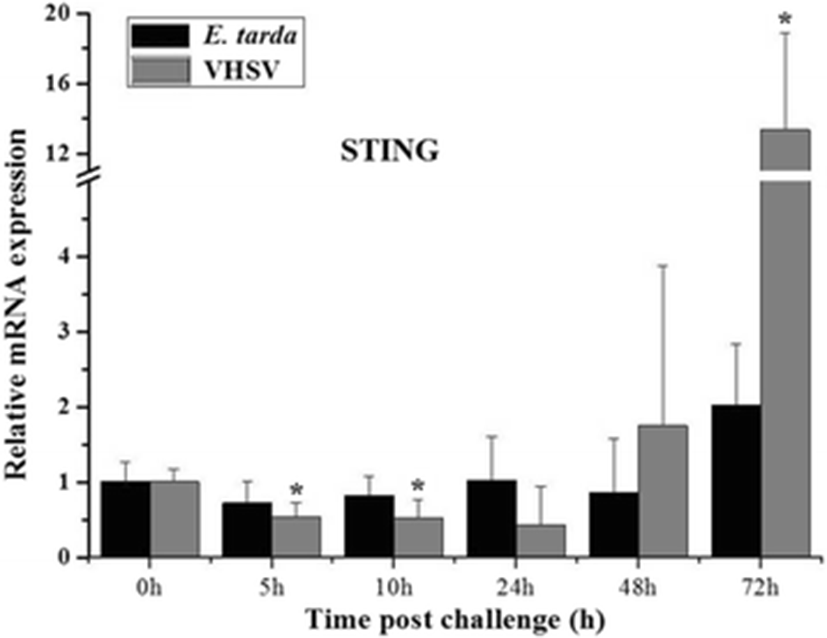
To understand the immune response of PoSTING, its temporal expression was assessed in the kidney following bacterial (E. tarda) and viral (VHSV) stimulation. The results revealed that there were no significant changes in the expression of PoSTING after E. tarda stimulation. Upon challenge with the virus, remarkably higher expression (over 13-fold) was detected at 72 h of injection compared to the expression in the un-injected control (0 h), whereas the expression was significantly downregulated at 5 and 10 h of injection (Fig. 5).
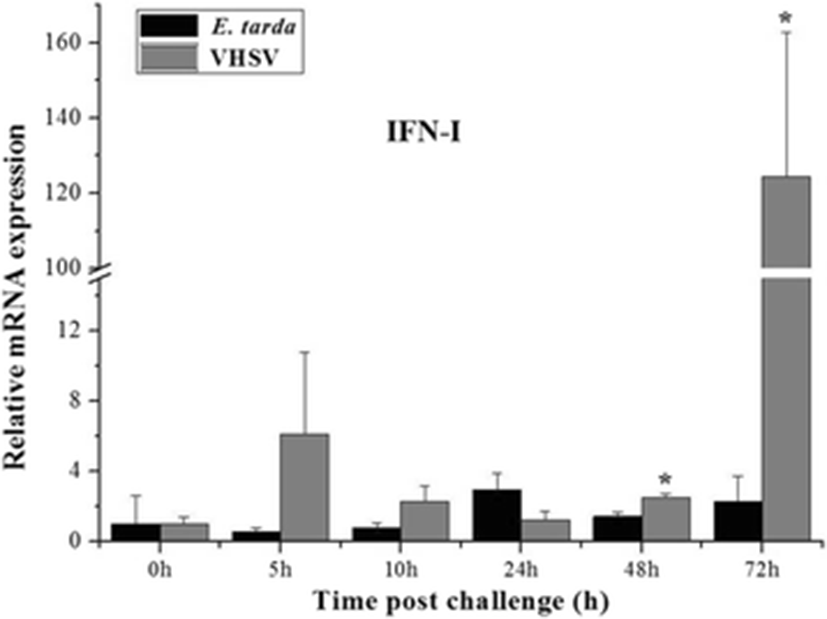
Similar expression patterns were observed for the expression of PoIFN-I after E. tarda and VHSV challenge. The expression of PoIFN-I was greatly elevated at 72 h of VHSV injection by 124-fold compared to the expression in that of un-injected control. Moreover, the expression was also significantly upregulated at 48 h of VHSV injection. The expression of PoIFN-I did not change with the bacterial challenge, as was observed for PoSTING (Fig. 6).
Discussion
The recognition of pathogenic microbes or microbial-derived elements is a vital immune process in the biological system that protects the organisms from invading pathogens. STING has been identified as an important adaptor protein that can recognize cytosolic nucleic acids (Abe et al. 2013). In this study, full-length cDNA of a STING gene was identified and characterized from the olive flounder. Bioinformatics analysis revealed that PoSTING contains three putative transmembrane (TM) domains. No signal peptide was detected in PoSTING by the SignalP program. However, some of previous studies have been reporting the existence of a signal sequence at the N-terminal region (Sun et al. 2011; Ge et al. 2015). Previous studies have reported that STING is a transmembrane protein located in the ER, and it facilitates the production of viral signaling molecules, such as Type I interferon (IFN) and interferon regulatory factor 3 (IRF3) (Ishikawa and Barber 2008; Zhong et al. 2008). It has also been demonstrated that the TM domains of STING are required to interact with the mitochondrial antiviral signaling protein (MAVS) in order to activate the IRF3 and induce the IFNs (Zhong et al. 2008). Moreover, the TM domains of STING are essential for its localization and oligomerization (Sun et al. 2009). Deletion of the TM domains alters the distribution of STING protein in the cells and abolishes its dimerization, which is important for its self-activation and subsequent downstream signaling (Sun et al. 2009). Thus, the TM domains in PoSTING protein might be involved in these kinds of activation related to the antiviral response. However, more studies are needed for understanding the real mechanisms.
The expression of STING genes have been examined in various tissues under normal physiological conditions. A previous study in mouse demonstrated that a high expression of STING was observed in the spleen and thymus, whereas moderate expression was observed in lung and kidney tissues (Sun et al. 2009). The ubiquitous expression of STING mRNA has been reported in the teleost, as well. In grass carp, the expression of STING mRNA was high in the foregut, skin, midgut, gill, and hindgut (Feng et al. 2014). The expression of STING mRNA was high in the gill, spleen, and brain tissues compared to that in the other tissues analyzed (Huang et al. 2015). In the present study, we observed high degree of PoSTING expression in the spleen, head kidney, and kidney, which are immune-related organs. A comparison of these results with those of previous studies suggests that the expression of STING might be species specific. However, in most of the species that were examined, higher expression levels were observed in the organs that are highly involved in immune regulations, implying the involvement of this protein in the process of immunity.
To understand the antimicrobial response of PoSTING, its expression patterns were examined in the kidney, which is the key organ central to several major biological systems, such as osmoregulation and immunity (Schmitz et al. 2016), upon bacterial and viral challenge. According to the qPCR results, significant modulations were detected only in the viral challenge experiment. Similarly, it was observed that the grass carp reovirus (GCRV) and Poly I:C trigger the expression of grass carp STING gene, whereas lipopolysaccharide (LPS; a bacterial component) stimulation had no effect on the expression. However, the expression of STING gene was significantly upregulated after peptidoglycan (a cell wall component of Gram-positive bacteria) stimulation in grass carp (Feng et al. 2014). In contrast, the STING expression was robustly upregulated in the spleen tissue by Singapore grouper iridovirus (SGIV), Poly I:C, and LPS stimulation (Huang et al. 2015). Taken together, these results indicate that the STING gene is mostly involved in the immune response against viral attacks.
To further understand the association of the STING gene with IFNs, we analyzed the expression of olive flounder IFN-I after bacterial and viral infection. The results showed similar patterns of expression of PoIFN-I and PoSTING transcripts. STING was recently found to be an essential adaptor to activate the retinoic acid-inducible gene I (RIG-I) and TANK-binding kinase 1 (TBK1) by initiating IFN expression, which might facilitate the immune responses against viral attack (Sun et al. 2011), and the overexpression of STING activated the transcription factors, NF-kB, and IRF3 and stimulated the IFN-I production (Konno et al. 2013). Another study reported that the STING protein in fish might assist the activation of IFN through IRF3 and IRF7 transcription (Sun et al. 2011). Together, these findings suggest that PoSTING plays a critical role in IFN-I induction and, thereby, triggers the cellular antiviral responses.
Conclusions
In summary, an ortholog of STING was identified from olive flounder and was characterized. Bioinformatics analysis revealed that PoSTING contained characteristic STING superfamily domain and three transmembrane domains as in the case of its counterparts in other species. The phylogenetic analysis showed distinct evolution of the teleost STING compared to those of other vertebrate species. Ubiquitous expression of PoSTING transcripts was detected in healthy fish, with the highest expression observed in the spleen tissue according in the qPCR analysis. Significantly upregulated expression of PoSTING mRNA was detected in kidney at 72 h of VHSV injection, whereas no change in the expression was observed upon bacterial stimulation. Similarly, VHSV infection triggered the PoIFN-I transcription at the same time point of the experiment, indicating the association of PoSTING with the antiviral response via PoIFN-I activation.