Background
The import value of live aquarium fish into South Korea has increased by more than twofold from $2.011 million in 1997 to $4.724 million in 2010 and continues to increase each year (Korea Agro-Fisheries & Food Trade Corporation in South Korea). Most of the ornamental fish (90%) imported into South Korea are freshwater species (about 1000 in total) that are cultured in aquariums. However, marine species (approximately 500) are caught in the wild. Most of the freshwater fish are imported from tropical Asian countries such as Malaysia, Indonesia, and China (Moravec et al. 1999), whereas marine fish are from Indonesia, the Philippines, and Singapore. As a result, the large number of imported fish may have exacerbated the problem of exotic fish pathogens being brought into South Korea.
Import of a variety of live fish species can lead to the unintended introduction of pathogens. In Australia, for example, the government has stringent regulations for live imported ornamental species that include mandatory quarantine periods of 1–3 weeks and health certification. Despite these measures, there have been many outbreaks of exotic viral, bacterial, fungal, protozoan, and metazoan infections among ornamental fish in Australian aquaculture farms. Aeromonas salmonicida is associated with ulcers in pearlscale goldfish (Carassius auratus), and Edwardsiella ictaluri with systemic disease in rosy barbs (Pethia conchonius) (Humphrey 1995). E. tarda was isolated from fighting fish (Betta splendens) imported from Singapore and quarantined in Australia in 1985 (Humphrey et al. 1986). Although E. tarda is a ubiquitous bacterium in freshwater environments, it was not known to cause mortality in aquafarm species and was found during a large survey of bacterial pathogens infecting salmonids in Australia between 1981 and 1985 (Humphrey et al. 1987). Furthermore, it was found to cause subdermal abscesses in wild eels (Anguilla reinhardtii) in Queensland in 1990 (Eaves et al. 1990) and mortality in farmed rainbow trout (Oncorhynchus mykiss) in New South Wales in 1993 (Reddacliff et al. 1996). Non-quarantined diseases are negligible. When however, a new or exotic disease is introduced, there is always the risk of serious pathogenic or transmissible diseases, resulting in enormous economic losses in commercial aquaculture or in wild aquatic systems.
The role of ornamental fish in the transmission of non-quarantine pathogens and the establishment of infection and disease has received little attention in South Korea. The Korean government has been investigating the influx of aquatic quarantine pathogens via imported living aquatic animals (National Fishery Products Quality Management Service in South Korea). However, these surveys had not focused on non-quarantine diseases associated with imported ornamental finfish until 2015.
We have addressed this in the current study by examining the presence of non-quarantine bacterial and viral pathogens in imported ornamental finfish, identification of the pathogens, and phylogenetic analysis of the diseases detected.
Methods
We examined the presence of microbial pathogens in ornamental finfish imported into Korea. Ornamental finfish specimens varied in terms of the number of samples and their size and weight (Table 1). Imported fish were randomly sampled for 6 months from February through July 2016. Fish sampled included ocellaris clownfish (Amphiprion ocellaris), pearlscale goldfish (Carassius auratus), and Nile tilapia (Oreochromis niloticus) at ten fish per month. Sutchi catfish (Pangasius sutchi) was imported every month with 20 fish being sampled each month. Five fish each of lemon damsel (Pomacentrus moluccensis), caerulean damsel (Pomacentrus caeruleus), and whitetail dascyllus (Dascyllus aruanus) along with three pearl-spot chromis (Chromis notata) were sampled in February and March, followed by ten fish each per month being sampled April through July. Five colored carp (Cyprinus carpio haematopterus) were sampled in February, and ten were sampled each month from March through July. Mandarin fish (Synchiropus splendidus) were sampled over 4 months from February through May, at a frequency of five fish in February and March and ten in April and May.
aMonthly sampling numbers for each species of fish from February to July 2016 are shown
Bacterial and viral pathogens in ornamental finfish were selected based on those most likely to infect finfish according to information from the World Organization for Animal Health (OIE) and survey data from the National Fishery Products Quality Management Service (NFQS) in South Korea for species imported in 2012–2015. Specifically, we selected pathogens for which there are standard international methods for detection (Table 2). Bacterial pathogens included Lactococcus garvieae, Yersinia ruckeri (the cause of enteric redmouth disease (ERM)), and the etiological agents of vibriosis, such as Vibrio anguillarum, V. alginolyticus, V. harveyi, V. vulnificus, V. penaeicida, and Photobacterium damselae (formerly V. damsel). Viral pathogens included European catfish virus (ECV), infectious hematopoietic necrosis virus (IHNV), viral nervous necrosis (VNN), and infectious spleen and kidney necrosis virus-like (ISKNV-like). Olive flounder (Paralichthys olivaceus) artificially infected with Vibrio anguillarum or viral hemorrhagic septicemia virus (VHSV) was used as positive experimental controls for each polymerase chain reaction (PCR) assay used for disease identification. The positive control for the PCR method was used to eliminate uncertainty regarding the method of disease detection in PCR assay.
|
Pathogen |
Primer name |
Sequence |
Reference |
|---|---|---|---|
|
L. garvieae |
pLG1 |
5′-CAT AAC AAT GAG AAT CGC-3′ |
Baeck et al. 2006 |
|
pLG2 |
5′-GCA CCC TCG CGG GTT G-3′ |
||
|
Y. ruckeri |
YER8 |
5′-GCG AGG AGG AAG GGT TAA GTG-3′ |
Gibello et al. 1999 |
|
YER10 |
5′-GAA GGC ACC AAG GCA TCT CTG-3′ |
||
|
V. anguillarum |
Van-ami8 F |
5′-ACA TCA TCC ATT TGT TAC-3′ |
Gonzalez et al. 2003 |
|
Van-ami417 R |
5′-CCT TAT CAC TAT CCA AAT TG-3′ |
||
|
V. alginolyticus |
gyrB Fw |
5′-GAG AAC CCG ACA GAA GCG AAG-3′ |
Zhou et al. 2007 |
|
gyrB Re |
5′-CCT AGT GCG GTG ATC AGT GTT G-3′ |
||
|
V. harveyi |
VH-4F |
5′-GTG ATG AAG AAG CTT ATC GCG ATT-3′ |
Kim et al. 2014 |
|
VH-7R |
5′-CGC CTT CTT CAG TTA ACG CAG G-3′ |
||
|
V. vulnificus |
vvhA Fw |
5′-TTC CAA CTT CAA ACC GAA CTA TGA-3′ |
Panicker et al. 2004 |
|
vvhA Re |
5′-ATT CCA GTC GAT GCG AAT ACG TTG-3′ |
||
|
V. penaeicida |
Fw |
5′-AA TAT TGC ACA ATG GGC GC-3′ |
Saulnier et al. 2000 |
|
Re |
5′-G CGC TTT ACG CCC AGT AAT TCC G-3′ |
||
|
P. damselae |
PD-2F |
5′-CAA GAC ATC ATC GAT GTG ATG CGT-3′ |
Kim et al. 2014 |
|
PD-2R |
5′-GAA ACT TTA CCA TCT ACC ACT TTG-3′ |
||
|
European catfish virus (ECV) |
Fw |
5′-ATGCGCTCCTCCGGGTCAAAG-3′ |
OIE 2011 |
|
Re |
5′-CATCATGAACGGCTCGATGG-3′ |
||
|
Infectious hematopoietic necrosis virus (IHNV) |
IHNV-GF |
5′-AGA GAT CCC TAC ACC AGA GAC-3′ |
Emmenegger et al. 2000 |
|
IHNV-GR |
5′-GGT GGT GTT GTT TCC GTG CAA-3′ |
||
|
Viral nervous necrosis (VNN) |
VNN F |
5′-CGT GTC AGT CAT GTG TCG CT-3′ |
Nishizawa et al. 1996 |
|
VNN R |
5′-CGA GTC AAC ACG GGT GAA GA-3′ |
||
|
Infectious spleen and kidney necrosis virus–like (ISKNV-like) |
P1 F |
5′-ATG TCT GCA ATC TCA GGT-3′ |
Xu et al. 2008 |
|
P2 R |
5′-TTA CAG GAT AGG GAA GCC TG-3′ |
||
|
NP1 (NP nested) |
5′-GCG TTT GAT GCG ATG GAG AC-3′ |
||
|
NP2 (NP nested) |
5′-ACG GCA GAG ACA CGG TAG GC-3′ |
To detect bacterial or viral infections in imported species, we analyzed quarantine records and procured regular samples from import companies listed in the NFQS. Samples were kept in their imported culture water, unexposed to the Korean environment. The finfish were anesthetized, their livers, spleens, and kidneys harvested using sterilized dissection tools, and the tissues evaluated for disease identification. As the first line of screening, we used thiosulphate-citrate-bile salts-sucrose (TCBS) agar plates as a selective medium for identifying Vibrio bacteria. To identify Vibrio spp., the yellow or green colonies on the TCBS medium were identified by PCR assay. As a large number of yellow or green colonies of different types formed, these were classified according to those with the same morphology, and 5–10 different types of colonies were used for PCR diagnosis.
For detection of the other pathogens (L. garvieae, Y. ruckeri, ECV, IHNV, VNN, and ISKNV-like), the collected target organs of the finfish were pooled into two or four groups with five fish per group, and the genomic DNA extracted using a GeneAll Exgene Tissue SV kit (GeneAll Biotechnology, Seoul, Korea), or the genomic RNA derived from virus purified using a RNeasy Mini kit (Qiagen, Valencia, CA, USA). The RNA was reverse transcribed to generate cDNA using Power cDNA synthesis kit (INtRON, Korea). The isolated genomic DNA and cDNA were used as template in PCR assays to detect the genes of the pathogen.
Pathogen detection and analysis were performed using PCR methods previously described, as referenced in Table 2. Details on the primer sets used for the detection of each pathogen are shown in Table 2. PCR mixes contained 10 μl of 2 × ExPrime Taq™ Premix (GENET BIO, Korea), which included 1 unit of ExPrime Taq DNA Polymerase, Tris-HCl (pH 9.0), PCR enhancer, (NH4)2SO4, 4 mM MgCl2, enzyme stabilizer, sediment, loading dye, and 2.0 mM dNTPs mixture; 0.5 μM of each primer of the relative primer sets; 1 μl of 10−1 diluted total DNA template; and distilled water to a final total volume of 20 μl. The thermal cycling profiles included an initial 95 °C for 3 min (tissue-extracted DNA) or 10 min (colony-extracted DNA), then 30 sequential amplification cycles, followed by a final extension step of 72 °C for 7 min. The amplification cycles were pathogenic-specific and consisted of denaturation at 95 °C for 30 s, annealing at 50 °C for 1 min, and template extension at 72 °C for 30 s for L. garvieae, ESV, and IHNV; denaturation at 95 °C for 30 s, annealing at 58 °C for 1 min, and template extension at 72 °C for 30 s for V. anguillarum, V. penaeicida, V. vulnificus, NNV, and ISKNV-like; or denaturation at 95 °C for 30 s, annealing at 60 °C for 1 min, and template extension at 72 °C for 30 s for Y. ruckeri, V. harveyi, V. alginolyticus, and P. damselae. The PCR products were sequenced by Cosmo Genetech (Seoul, Korea) and confirmed using the basic local alignment search tool (BLAST) on the National Center for Biotechnology Information (NCBI) website (https://www.ncbi.nlm.nih.gov).
For the bacterial pathogens, 16S rRNA gene sequences were obtained for L. garvieae, P. damselae, and Y. ruckeri. The gyrase B subunit (gyrB) gene sequence was obtained for V. alginolyticus. For the Vibrio spp., amiB sequences of V. anguillarum, which encode N-acetylmuramoyl-L-alanine amidase, RNA polymerase beta subunit (rpoB) sequences of V. harveyi and vvhB sequences of V. vulnificus, which encodes a chaperone, were obtained from analysis of the PCR products. For analysis of the viral pathogen ECV, gene sequences of the major capsid protein (MCP) were obtained. Each sequence detected was aligned using the NCBI GenBank database.
For phylogenetic analysis, the aligned sequences were evaluated using the MEGA 7 (Molecular Evolutionary Genetic Analysis 7) software program (Kumar et al. 2016). The alignments of the predicted protein or nucleotide sequences were revised by excluding regions with uncertain alignment at the ends of the sequences. The phylogenetic trees were constructed using the neighbor-joining method with bootstrap analysis (1000 replications), which showed that the pathogen genes detected in this study were placed into the same clade with respective species in GenBank.
Results
We analyzed the monthly prevalence of bacterial and viral pathogens based on the total finfish samples (Table 3). Two pathogens were detected in February, V. anguillarum in a pearl-spot chromis and V. harveyi in an ocellaris clownfish. No pathogens were detected in the imported finfish tested in March. Among the fish species imported in April, there were two cases of V. harveyi detected in mandarin fish and one case of V. vulnificus detected in each of the pearl-spot chromis and caerulean damsel fish. In May, there was a positive finding for L. garvieae in a group of sutchi catfish, V. vulnificus in an ocellaris clownfish, and two cases of ECV detected in separate groups of sutchi catfish. In June, there was one case each of Y. ruckeri in a group of pearlscale goldfish, P. damselae in an ocellaris clownfish, and V. alginolyticus in a lemon damsel. In July, eight specimens were tested positive for genetic material from bacterial pathogens. These included three cases of vibriosis (V. alginolyticus in a lemon damsel, V. vulnificus in both a lemon damsel and a caerulean damsel, and P. damselae in a caerulean damsel); a case of Y. ruckeri in a pearlscale goldfish and two cases in separate groups of colored carp; and one case of L. garvieae in a lemon damsel.
Phylogenetic trees based on the detected genes of the pathogens were generated using MEGA 7 software. Alignment with known sequences in the GenBank database showed that the genetic sequence detected in lemon damsel, which encoded for only 385-aa, had a high similarity (99%) with the 16S rRNA genes of L. garvieae from Oncorhynchus mykiss (KM604702.1) in India and Oreochromis niloticus (KM209202.1) in Indonesia. The 388-aa fragment detected in ocellaris clownfish showed 99% similarity with GenBank sequences for the 16S rRNA gene of P. damselae from Lates calcarifer in India (MF164181.1) and 96% similarity with Oplegnathus fasciatus of China (KF956381.1) (Fig. 1a). The genetic sequence derived from the pathogen isolated from the colored carp encoded for a 496-aa fragment and shared 99% similarity with the 16S rRNA gene of Y. ruckeri from Salmo salar of Norway (CP023184.1) and grass carp of China (KF003196.1) (Fig. 1b).
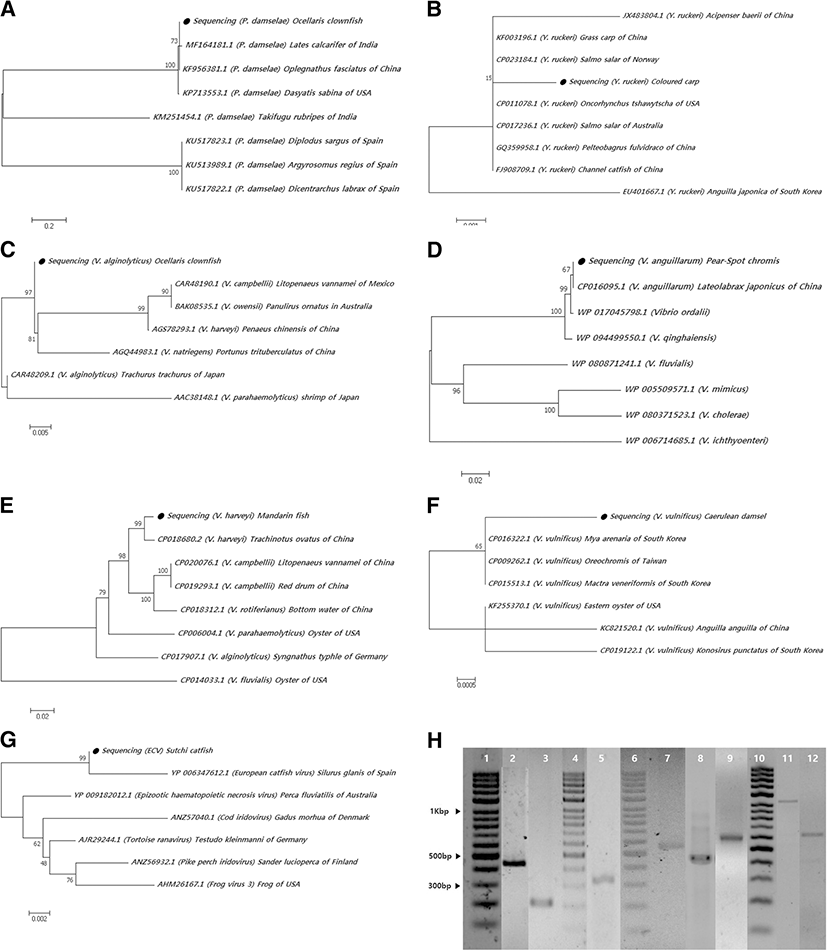
In the Vibrio spp., the 349-aa gyrB sequence isolated from the lemon damsel showed relatedness to V. alginolyticus from Trachurus trachurus in Japan (CAR48209.1) with a 97% sequence similarity (Fig. 1c). The 551-aa fragment of the V. anguillarum amiB that was detected in pearl-spot chromis had 99% similarity to the amiB of V. anguillarum isolated from Lateolabrax japonicus of China (CP016095.1) (Fig. 1d). The 481-aa region of rpoB detected in mandarin fish demonstrated 99% identity to V. harveyi rpoB from Trachinotus ovatus of China (CP018680.2) (Fig. 1e). The 579-aa segment of V. vulnificus vvhB detected in caerulean damsel was 94% similar to V. vulnificus vvhB from Mya arenaria (CP016322.1), Mactra veneriformis (CP015513.1) of South Korea, Oreochromis of Taiwan (CP009262.1), and eastern oyster of the USA (KF255370.1) (Fig. 1f).
In addition to the bacterial sequences detected, the viral sequences also aligned with known sequences in the GenBank database. The 395-aa fragment of the ECV MCP gene detected from sutchi catfish demonstrated 99% sequence similarity with the ECV gene of Silurus glanis of Spain (YP 006347612.1) (Fig. 1g).
Discussion
The occurrence of bacterial or viral diseases in aquaculture is a major problem that can lead to enormous economic losses worldwide. Vibrio species are widely distributed in marine environments and estuaries and are some of the most abundant pathogens in aquafarms (Austin and Austin 1993a; Bergh et al. 2001; Hjeltnes and Roberts 1993; Lightner 1993). V. anguillarum, V. alginolyticus, V. harveyi, and V. vulnificus are among the main pathogens causing vibriosis in several finfish species (Austin and Zhang 2006; Balebona et al. 1998b; Romalde 2002), including in South Korea.
We detected the V. alginolyticus gyrB gene in lemon damsel during a 2-month period (detection rates of 10% in both June and July). The gyrB gene detected in caerulean damsel had 97% similarity with the V. alginolyticus isolate from Trachurus in Japan according to sequence alignment and analysis, with a close association being demonstrated in the phylogenetic tree. V. alginolyticus is responsible for epizootic outbreaks and mortality in farmed gilt-head sea bream (Sparus aurata L.) and puffer fish (Fugu vermicularis vermicularis) (Balebona et al. 1998b; Colorni et al. 1981; Noguchi et al. 1987) and has caused massive losses for the aquaculture industry in China (Xie et al. 2005). In sea bream, symptoms of infection include septicemia, hemorrhage, and fluid accumulation in the peritoneal cavity (Balebona et al. 1998a; Colorni et al. 1981). Ours is the first report of V. alginolyticus infection in lemon damsel.
V. vulnificus was detected in lemon damsel in July, caerulean damsel in April and July, pearl-spot chromis in April, and ocellaris clownfish in May, at a 10% detection rate in each case. Phylogenetic analysis revealed that the vvhB detected in caerulean damsel shared 94% similarity with the V. vulnificus isolates from Mya arenaria and Mactra veneriformis of South Korea. This pathogen has been shown to cause infectious diseases in Japanese and European eels in Spain, Sweden, and Norway (Biosca et al. 1991, 1997; Muroga et al. 1976). V. vulnificus has been detected in marine fish collected along the west coast of India at an incidence of 16.6% (Thampuran and Surendran 1998) and has also been detected in other countries such as Spain, the Netherlands, and Japan.
V. anguillarum was detected in approximately 1/3 of pearl-spot chromis imported in February. The amiB of V. anguillarum that was detected from pearl-spot chromis had 99% similarity with the amiB of V. anguillarum derived from Lateolabrax japonicus of China. V. anguillarum was first reported in European eels (Bergman 1909) and is widely found in both cultured and wild fish, including Salmo salar L. (salmon), rainbow trout (Oncorhynchus mykiss), bream, eel, mullet, catfish, and tilapia (Oreochromis spp.) in salt or brackish water. Infections cause fatal hemorrhagic septicemic disease (Aguirre-Guzman et al. 2004; Frans et al. 2011; Paillard et al. 2004; Toranzo et al. 2005).
The V. harveyi rpoB sequence was detected in ocellaris clownfish (10% infection rate in February) and mandarin fish (20% infection rate in April). The rpoB gene sequence detected in the mandarin fish demonstrated 99% identity with the gene from Trachinotus ovatus of China. This pathogen is known to affect crevalle jack (Caranx hippos), summer flounder (Paralichthys dentatus), sandbar shark (Carcharhinus plumbeus), and lemon shark (Negaprion brevirostris) (Austin and Zhang 2006; Kraxberger-Beatty et al. 1990; Bertone et al. 1996; Colwell and Grimes 1984; Grimes et al. 1984; Lee et al. 2002; Soffientino et al. 1999).
P. damselae was detected in ocellaris clownfish (10% detection rate in June) and in caerulean damsel (10% detection rate in July). Analysis of P. damselae using the 16S rRNA gene detected in ocellaris clownfish revealed that it had 96% similarity to the P. damselae derived from Oplegnathus fasciatus of China and resulted in the relatedness demonstrated by the phylogenetic tree. This pathogen has previously been shown to affect cultures of red-banded sea bream, common sea bream, white seabream, and marine rainbow trout in Denmark (Labella et al. 2011; Pedersen et al. 2009). The mortality rates range between 5% and 94% depending on the season, and the species and age of the fish (Labella et al. 2011).
L. garvieae was detected in lemon damsel (50% infection rate in July) and sutchi catfish (25% infection rate in May). Sequencing analysis of the 16S rRNA gene detected in lemon damsel had high similarity (99%) with the L. garvieae 16S rRNA gene isolated from Oncorhynchus mykiss in India and Oreochromis niloticus in Indonesia. This pathogen was first discovered in rainbow trout raised on a Japanese fish farm in the 1950s (Hoshina et al. 1958) but has since been isolated as a disease-causing agent in rainbow trout, yellowtail, tilapia, Japanese eel (Anguilla japonica), olive flounder (Paralichthys olivaceus), gray mullet catfish, wild wrasse (Coris aygula), black rockfish (Sebastes schlegeli), amberjack (Seriola dumerili), and kingfish (Seriola lalandi) (Chen et al. 2001; Colorni et al. 2003; Fadaeifard et al. 2012; Kang et al. 2004; Kawanishi et al. 2005; Kusuda et al. 1991; Lee et al. 2001; Prieta 1993; Ravelo et al. 2003). Various countries with aquafarms have reported L. garvieae infections, including Israel, Italy, Taiwan, Spain, Japan, Australia, Turkey, and the UK (Baya et al. 1990; Eldar et al. 1994, 1995, 1996; Kitao 1993; Kusuda et al. 1976; Nieto et al. 1995; Rasheed and Plumb 1984), as well as South Korea (Kang et al. 2004).
Y. ruckeri was detected in pearlscale goldfish (one of two groups in June and July each) and colored carp (both groups in July). The Y. ruckeri 16S rRNA gene derived from colored carp showed 99% similarity with the Y. ruckeri 16S rRNA gene isolated from Salmo salar of Norway and grass carp of China. The disease ERM, which is caused by Y. ruckeri, can lead to mortality in farmed salmonids (Austin and Austin 1993b). There have been no reports to date of ERM in pearlscale goldfish or colored carp; in addition, it has only been associated with farmed eel in South Korea (Joh et al. 2010, 2013).
Finally, ECV was the only viral pathogen detected in the current study but was found at a rate of 10% in sutchi catfish imported in May. The ECV MCP gene detected in sutchi catfish showed 99% sequence similarity with that isolated from Silurus glanis of Spain. In Germany, ECV caused high morbidity and mortality in sheatfish and catfish hosts in a 1989 outbreak, causing 100% mortality in infected fish (Ahne et al. 1989).
In this study, Vibrio species, L. garvieae and P. damselae, were already known to exist in South Korea, but ECV and Y. ruckeri have not been reported in South Korea yet. Therefore, Y. ruckeri and ECV are exotic infectious diseases, and it is necessary to continuously monitor and analyze the effects of the pathogenicity of these diseases.
We were unable to identify any individuals in the imported samples with unusual symptoms of disease infection. The culture water imported with the samples was cloudy only for the lemon damsel and ocellaris clownfish. However, we did not detect pathogens in culture water. We did not observe any symptoms of latent stage diseases in the ornamental finfish. We also screened for genes of V. penaeicida, IHN, VNN, and ISKNV-like as pathogens in imported finfish, but none were detected during the sampling period of the study. Furthermore, we did not detect genes of the selected pathogens in whitetail dascyllus or Nile tilapia during the test period.
Conclusions
The frequency of detecting bacterial and viral pathogens among the finfish imported into South Korea from February through July 2016 gradually increased over time. This may have been associated with an increase in water temperature. Domestic interest in imported aquatic species is increasing; exotic finfish are bred as a hobby, as well as for consumption. However, this may lead to an increase in imported pathogens in South Korea. In Australia, a wide range of viruses, bacteria, and parasites was identified in ornamental fish during quarantine in the period from 1999 to 2004 (Chong and Whittington 2005), and these pathogens had negative effects in aquatic animals. It is therefore essential to exercise greater caution during importation and to engage in more careful monitoring of infectious diseases in South Korea to protect indigenous aquatic species. This study was the first to identify the species and origin of the detected pathogens and the presence or absence of pathogens (non-quarantine diseases) in imported ornamental finfish. Future studies will investigate pathogenicity in finfish using stock cultures of the pathogens responsible for the diseases that were detected.