Introduction
Of more than 140 enteric viruses identified from human faces, NoV has been the leading causative pathogen of acute viral gastroenteritis (Leclerc, et al., 2000). NoV is positive-sense single stranded RNA virus classified into family Caliciviridae and comprise six genogroups (Trujillo et al., 2006). GI and GII groups are more commonly associated with human illness (Zheng et al., 2006) and each group is currently subdivided into 9 and 22 genotypes, respectively (Lu et al., 2015). NoV transmission pathways are the fecal-oral route by consumption of uncooked contaminated food as well as by contact of the person, aerosol and the solid surface (Cho et al., 2011).
NoV is the common cause of gastrointestinal illness all over the world (de Wit MA et al., 2001; Patel et al., 2008; Scallan et al., 2011). Many countries reported abundant cases of NoV infection with very significant economic costs on a regular basis. In the United States, NoV is estimated to cause 21 million cases (Scallan et al., 2011) and $184 million of the cost of illness annually (Batz et al., 2011). In Korea, NoV infection was first officially reported in 1999. In 2006, the outbreak occurred in which 2,400 students were infected through consumption of contaminated food via groundwater at the food service centers of 31 schools, which aroused social concern about the NoV infectious disease (Lee et al., 2013).
There have been many cases of NoV infection associated with the consumption of contaminated shellfish (Koo et al., 2016; Prato et al., 2004; Smith et al., 2009; Webby et al., 2007; Westrell et al., 2010). Historically, the monitoring of regular fecal indicator bacteria in the shellfish production areas is the most common strategy to secure the safety of shellfish (Oh et al., 2015). However, evidences have demonstrated that NoV persists for longer periods than fecal indicator bacteria in actively filtering shellfish (Oh et al., 2015; Schwab et al., 1998). Major NoV contamination sources in shellfish production areas have been the discharges of untreated sewage from small individual septic tanks (Borchardt et al., 2011; Rodriguez et al., 2012).
Direct discharges into production areas have clearly shown the links between NoV in sewage and gastrointestinal illness (Nenonen et al., 2008; Ueki et al., 2005; Wall et al., 2011). In other words, raw sewage may act as a valuable source to characterize the prevalence of enteric viruses in a human population. Accordingly, several studies have used raw sewage for molecular epidemiology as a source of information on the viruses circulation through the fecal-oral route (Fumian et al., 2019; La Rosa et al., 2017; Montazeri et al., 2015; Nenonen et al., 2008; Prado et al., 2018).
In this study, we examined the prevalence of NoV in sewage samples from the drainage basin of Jaran Bay, Korea. We assessed genotypes and diversity of NoV strains detected in sewage samples collected from individual septic tanks in January, May and July of 2017.
Materials and Methods
Sampling was conducted at 7 villages along the coastline of Jaran Bay facing Goseong County and Tongyeong, South Korea (Fig. 1). In each village, five septic tanks located closest to the coast were selected for sampling. Jaran Bay is surrounded by the coastline of Samsan-myeon and Hail-myeon of Goseong County and the southern part is open to the Pacific Ocean. About 800 shellfish farming licenses were issued in about 76 km2 coastal area.
In January, May, and July of 2017, 1 L of sewage was collected from each septic tank and transported to the laboratory at below 4°C and NoV genotype distribution was investigated among a total of 100 sewage samples. On the sampling day, 45 mL of sewage sample was centrifuged (170,000×g, 60 min, 4°C) using the ultracentrifuger (Beckman Coulter, Brea, CA, USA). The pellet was resuspended in 5 mL of glycine buffer (0.75 M glycine/0.15 M NaCl, pH 7.6) (Sigma-Aldrich, St. Louis, MO, USA) then 50 mL of 2× PBS (Merck, Darmstadt, Germany) as added. The solution was stirred for 20 min on ice and then centrifuged (12,000×g, 20 min, 4°C). The supernatant was adjusted to 45 mL using 1× PBS and then flocs were sedimented by centrifugation (170,000×g, 60 min, 4°C). The pellet was resuspended in 1 mL of 1x PBS and stored at –80 °C for downstream analysis.
Viral RNA was extracted from 500 μL of viral suspension using eMAG™ instruments (bioMerieux, Grenoble, France) according to the procedure of the eMAG-specific program (IND_B20t_On_Ves_SB_1). For detection of NoV from stool samples, RT semi-nested PCR was performed using a previously described primer sets (Table 1) (Cheon et al., 2010). Extracted RNA was firstly amplified using Verso 1-Step RT-PCR Kit ReddyMix (Thermo Fisher Scientific, Waltham, MA, USA) under the following conditions: a reverse transcription step at 45°C for 30 min and PCR involving an initial activation step at 94°C for 5 min, followed by 35 cycles each of 94°C for 30 sec, 55°C for 30 sec, and 72°C for 1 min 30 sec, with a final extension step at 72°C for 7 min. A semi-nested PCR amplification procedure was then performed using Top DNA Polymerase (Bioneer, Daejeon, Korea) as follows: 94°C for 5 min, followed by 25 cycles of 94°C for 30 sec, 55°C for 30 sec, and 72°C for 90 sec, followed by a final extension step at 72°C for 7 min. The amplified fragments were purified using the Wizard® SV Gel and PCR Clean-Up System (Promega, Madison, WI, USA) and then cloned using the TOPclonerTM TA core Kit (Enzynomics, Daejeon, Korea) according to manufacturer’s instruction. Plasmid DNA was purified using the AccuPrep® Plasmid Mini Extraction Kit (Bioneer), and DNA was sequenced by the Cosmogentech, Ltd., South Korea.
The analyzed nucleoprotein (N) site sequence of the isolated strain was identified using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and the genotype showing the highest homology through comparative analysis with the existing NoV standard strain sequence registered in GenBank (http://www.ncbi.nlm.nih.gov). Sequences were aligned using the ClustalW program, and genetic distances and phylogenetic trees between base sequences were inferred by the neighbor-joining method using the MEGA-X v 10.1.8 (Molecular Evolutionary Genetics Analysis version 10.1.8) program. Also, the bootstrap value was deduced from 1,000 resampled data.
Results
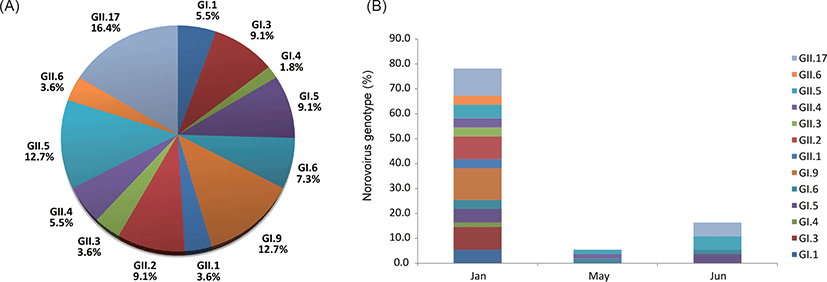
In January, May and July of 2017, a total of 100 raw sewage samples were collected from 35 septic tanks in 7 villages. In total, NoV GI or GII were detected in 22 (22.0%) and 24 (24.0%) samples, respectively. Ten samples contained both NoV GI and NoV GII (Table 2). The 22 GI-positive samples were genotyped as GI.1, GI.3, GI.4, GI.5, GI.6 and GI.9 and 24 GII positive samples were GII.1, GII.2, GII.3, GII.4, GII.5, GII.6 and GII.17. The genotypic distribution of the 13 NoV strains was as follows: GI.1 (5.5%, n=3); GI.3 (9.1%, n=5); GI.4 (1.8%, n=1); GI.5 (9.1%, n=5); GI.6 (7.3%, n=4); GI.9 (12.7%, n=7); GII.1 (3.6%, n=2); GII.2 (9.1%, n=5); GII.3 (3.6%, n=2); GII.4 (5.5%, n=3); GII.5 (12.7%, n=7); GII.6 (3.6%, n=2); GII.17 (16.4%, n=9) (Table 2). Among identified thirteen NoV genotypes, three genotypes (GI.9, GII.5 and GII.17) were dominant (Fig. 2A). NoV GII.17 showed relatively higher prevalence during the survey period. NoVs were detected in raw sewage samples collected at 23 out of 35 septic tanks in January (the winter of Korea), recording an exceptionally high overall detection rate of 65.7%. All thirteen genotypes were identified in January while three and four genotypes were identified in May and July, respectively (Fig. 2B).
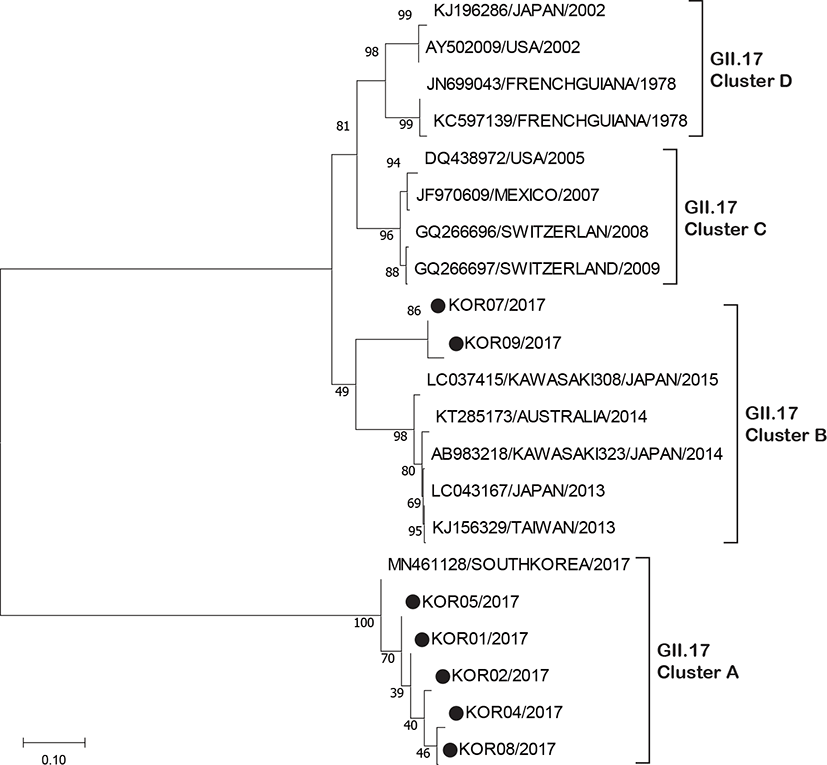
By phylogenetic analysis, the genetic diversity of NoV GII.17 strains detected with those of reference strains was evaluated using their partial capsid sequence. In the phylogenetic tree, 7 strains were clustered into two sub-clusters (Fig. 3). Five strains isolated clustered with recombinant type variant newly emerged in 2017 in South Korea (Cluster A) and 2 strains were clustered into the recently emerged NoV GII.17 Kawasaki variant (Cluster B).
Discussion
Although the investigation period was short, NoV detection rate and genotype diversity in this study showed remarkable seasonality. Ahmed et al. (2013) conducted a systematic review of report on monthly counts of NoV between 1997 and 2011 and analyzed seasonal patterns of NoV disease. The research identified that 41.2% of outbreaks and 52.7% of cases occurred in cold months. Koo et al. (2016) investigated molecular epidemiology of NoV infections from outbreaks in Busan, Korea from 2012 to 2015 and it was found that NoVs were most frequently detected in November (25.4%) and February (28.2%). A meta-analysis of NoV identified in raw sewage from various temperate climate countries showed a high degree of a winter peak for both GI and GII genogroups (Eftim et al., 2017). Our findings are broadly consistent with the NoV concept of a winter virus, at least in temperate climate. However, there is no opportunity to access sufficient reports of NoV outbreaks in Africa, South America, and tropical regions, so it is insufficient to assert the seasonality of NoV incidences and relevant acute gastroenteritis outbreaks.
Among identified thirteen NoV genotypes, NoV GII.17 showed relatively higher prevalence during the survey period. NoV GII.17 is an uncommon genotype which recently emerged in China and spread throughout other south-east Asian countries resulting in an increased number of acute gastroenteritis outbreaks reported within this region (Lu et al., 2015; Matsushima et al., 2015). The GII.17 Kawasaki 2014 has also been detected in many countries in North and South America, Europe and Oceania (Andrade et al., 2017; Lun et al., 2018; Medici et al., 2014; Oh et al., 2015; Parra & Green, 2015). The molecular epidemiology of acute gastroenteritis outbreaks, which have been mainly caused by NoV GII.4 variants since the mid-1990s, are changing globally with the emergence of NoV GII.17. Nam et al. (2014) reported that 35 acute gastroenteritis outbreaks occurred in Gyeonggi Province of South Korea in early 2015 were caused by NoV GII.17 Kawasaki 2014. Above findings encourage some authors to suggest GII.17 might replace GII.4 as the predominant NoV strain around world at some point (Bruggink et al., 2016).
In Brazil, it was presumed that GII.17 Kawasaki variant was introduced in 2014 during the 2014 FIFA World Cup event (Andrade et al., 2017). However, the molecular analysis of NoVs detected in raw sewage showed that this variant has already been circulating since mid-2013 in Brazil (Fumian et al., 2019). This could be related to asymptomatic or mild infections of this genotype that do not lead to hospital visits. So, the unmatched results may be a reflection of underestimation over circulation of NoV GII.17. Sewage analysis is an important molecular surveillance tool to understand the genetic diversity of enteric viruses in a human population (La Rosa et al., 2017; Mabasa et al., 2015; Montazeri et al., 2015; Prado et al., 2018). WHO used sewage to monitor poliovirus circulation in communities worldwide as a part of the Global Polio Eradication Initiative with combining clinical surveillance (Benschop et al., 2005).
If human enteric viruses including NoVs and hepatitis A virus are presenting in the stools originated relevant patients, surface water and sea water may be polluted with these viruses (Lodder et al., 2005; Metcalf et al., 1982). Shellfish accumulates NoV from surrounding water during filter-feeding in their digestive glands and other tissues within 4 to 24 h (Wang et al., 2008; Asahina et al., 2009), which may cause the gastrointestinal disease. Monitoring on oyster production areas in the United Kingdom, France and Ireland, NoV (GI plus GII) ranging from 102 genome copies/g to 104 genome copies/g was detected (Wyn-Jones & Sellwood, 2001). Monitoring on farmed and wild mussels, clams, and cockles detected NoV ranging from 101 to 104 copies/g in Galicia, Spain (Vilariño et al., 2009). Although various shellfish are almost consumed after heating and boiling but the heat treatment at the level of shells opening is insufficient to inactivate viruses (Myrmel et al., 2004).
Effective sewage treatment process is essential to prevent environmental contamination before the discharge of effluents into water bodies used by the public thus mitigating the risk of viral transmission to the population. However NoV is removed to only a limited extent by sewage treatment processes and, consequently, the virus is commonly detected in final discharges of waste water treatment plants under normal operational status (Carlos & Lees, 2014). The secondary strategy for the risk reduction is therefore to secure the prohibited zone (i.e. buffer zone) for sufficient dilution and dispersion of viral particles between the final discharge points of waste water treatment plants and the impacted seafood production areas.
In conclusion, we successfully identified the circulation of three major prevalent genotypes (GI.9, GII.5 and GII.17) in raw sewage samples during the study period. Among those, NoV GII.17 strains were considered emergent epidemic variants with widespread circulation. NoV showed a genetic diversity in raw sewage with thirteen major and minor genotypes. NoV surveillance strategy should include both environmental (sewage) and clinical data to reveal minor NoV genotypes likely cause asymptomatic or underreported infections in the local population. The strategy for mitigation of viral infection risk is the effective sewage treatment and sufficient dilution of final effluents before reaching to the seafood production areas.
