Introduction
Myxozoans are microscopic endoparasites that belong to the phylum Cnidaria; they complete their life cycles in vertebrate (intermediate hosts; mainly fish) and invertebrate (definitive hosts; annelids or bryozoans) hosts (Chang et al., 2015; Lom & Dyková, 2006). Myxozoans comprises more than 2,000 described species, several of which can cause diseases in economically important fish (Lom & Dyková, 2006). Notably, emaciation disease caused by myxosporean infection (Enteromyxum leei, Enteromyxum scophthalmi, and Sphaerospora fugu) causes mass mortality in important aquaculture fish worldwide (such as olive flounder Paralichthys olivaceus, gilt-head sea bream Sparus aurata, tiger puffer Takifugu rubripes, and turbot Scophthalmus maximus) (Diamant, 1992; Ogawa & Yokoyama, 2001; Palenzuela et al., 2002; Sekiya et al., 2016; Tun et al., 2002).
The starry flounder Platichthys stellatus of the family Pleuronectidae inhabits the coastal waters of the Pacific and Arctic oceans, and the East Sea of Korea (Kang et al., 2012; Orcutt, 1950). Since the inception of aquaculture of starry flounder began in the early 2000s, it has become a candidate marine flatfish for aquaculture in Korea as it can feed at low temperatures and is disease-resistant (Han et al., 2019; Kim et al., 2019; NIFS, 2019). Moreover, some fish farms in Jeju have attempted to culture starry flounder instead of olive flounder as the mortality of cultured olive flounder by emaciation disease has increased recently (Shim et al., 2019). The Marine Science Research Institute of Jeju National University monitored the parasitic infections of olive flounder and starry flounder that are cultured on Jeju island and found Enteromyxum species in starry flounder intestines. The present study aimed to identify Enteromyxum species and determine the effect of the parasitic infection of these species in cultured starry flounder.
Materials and Methods
Ten starry flounder individuals (n = 20, total length: 22.9 ± 2.0 cm; body weight: 183.8 ± 79.4 g) were obtained from a starry flounder farm on September 16 and November 10, 2022, respectively. Upon arrival to Marine Science Research Institute, the 20 individuals were administered at lethal over dose of Tricaine Methanesulfonate (Syndel, Ferndale, WA, USA), and wet smears of fresh gill, skin, and intestinal scrapings were examined using a light microscope. The kidney and liver tissues were used to isolate the bacteria, which were then incubated on a brain-heart infusion agar plate (BHIA) supplemented with 1% NaCl at 25°C for 48 h.
Twenty samples of 50–100 mg of intestinal tissue were collected in microtubes and stored at −20°C. DNA was extracted directly from the samples using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol and was eluted in 100 μL AE buffer. We amplified parts of the 18S rDNA by polymerase chain reaction (PCR) using a combination of primers (Table 1) designed in previous studies (Barta et al., 1997; Hillis & Dixon, 1991; Sekiya et al., 2016). In addition, E. leei was detected by PCR using primers specific for E. leei (Sekiya et al., 2016) (Table 1). The PCR comprised the following steps: initial denaturation at 95°C for 5 min, followed by 35 cycles at 95°C for 30 s, 58°C for 30 s, and 72°C for 30 s, and a final extension at 72°C for 7 min. PCR products were treated with the AccuPrep Genomic PCR Purification Kit (Bioneer, Daejeon, Korea) to remove excess primers and dNTPs and directly sequenced using BigDyeTM Terminator v3.1 in an ABI 3730xl Sequencer. BLASTN analysis was performed using National Center for Biotechnology Information database based on the 18S rDNA sequence of the isolate.
| Purpose | Target | Name | Sequence (5’-3’) | Forward/Reverse | References |
|---|---|---|---|---|---|
| Identification | 18S rRNA | 18e | CTGGTTGATCCTGCCAGT | Forward | Hillis & Dixon, 1991 |
| 18S rRNA | EL-F | GGATATTGTCAGAGTATGTT | Forward | Sekiya et al., 2016 | |
| 18S rRNA | mEL-R | AGAAGCCAACGTATATGATTA | Reverse | Sekiya et al., 2016 | |
| 18S rRNA | ERIB10 | CTTCCGCAGGTTCACCTA | Reverse | Barta et al., 1997 | |
| Quantitative PCR | 28S rRNA | EL-qF | ACTCGGCTATGTGGGCAGTG | Forward | Sohn et al., 2021 |
| 28S rRNA | EL-qR | ATTGTTAATTGCTAGTCGTAAAGAGCAAG | Reverse | Sohn et al., 2021 |
The total length and body weight of the fish were measured to calculate the condition factor (CF = body weight × 100 / [total length]3). To investigate the effect of the intensity of E. leei infection on the CF of starry flounders, quantitative PCR (qPCR) was carried out as the previously described (Sohn et al., 2021). The qPCR was performed in a 10 μL total reaction mixture containing 3 μL of template, 0.5 μL of each primer (Table 1) (10 pmol/μL), 5 μL of 2 × TaKaRa Ex Taq™ SYBR Premix (TaKaRa, Shiga, Japan) and 1 μL of nuclease-free water. The thermal profile was as follows: 95°C for 30 s, followed by 45 cycles at 95°C for 5 s and 60°C for 30 s.
Statistical analyses were performed using the SPSS 16.0 software (version 16.0; SPSS, Chicago, IL, USA). The relationship between CF of starry flounder and the intensity of E. leei infection were investigated using regression analysis and one-way analysis of variance. Differences were considered statistically significant at p < 0.05.
Results
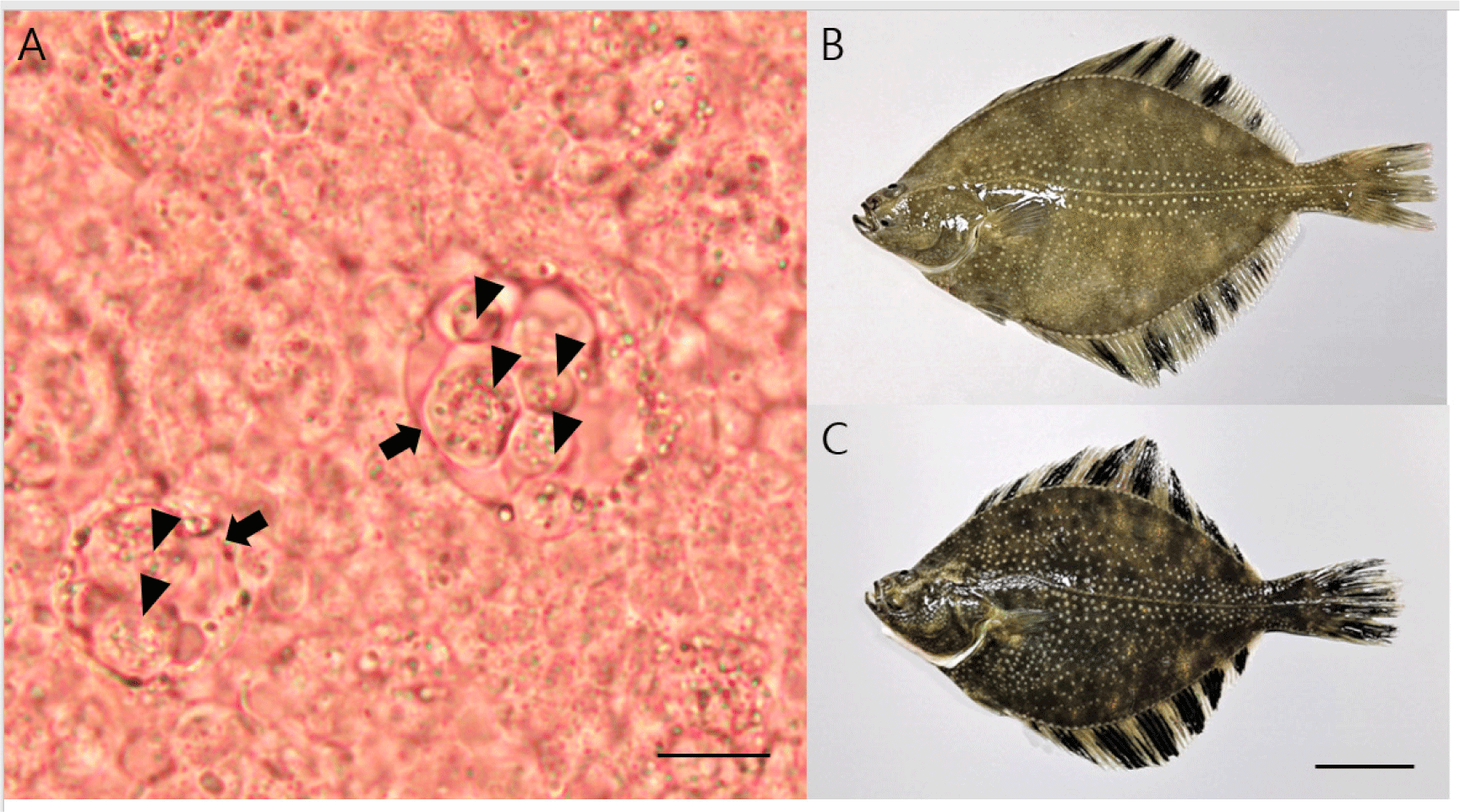
No parasitic infection was observed in the skin and gill samples; however, many primary cells (including accompanying cells) similar to the developmental stage of E. leei were found in the intestine samples (Fig. 1A). All the fish examined had no gross lesions, regardless of whether parasites were detected (Fig. 1B and 1C). Microscopic investigation showed that the detection rate for Enteromyxum species in the intestinal samples was 20% and 70% for samples obtained in September and November, respectively. Partial sequences of 18S rDNA (1,680 bp) were obtained from Enteromyxum species and deposited in GenBank (accession number OQ130171). The sequences showed 100% (1,680/1,680) nucleotide identity with the 18S rDNA (accession number MF161396) of E. leei. The PCR results (with primers specific for E. leei) showed 50% and 100% detection in September and November, respectively. No bacteria grew on BHIA plates from the kidney and liver specimens.
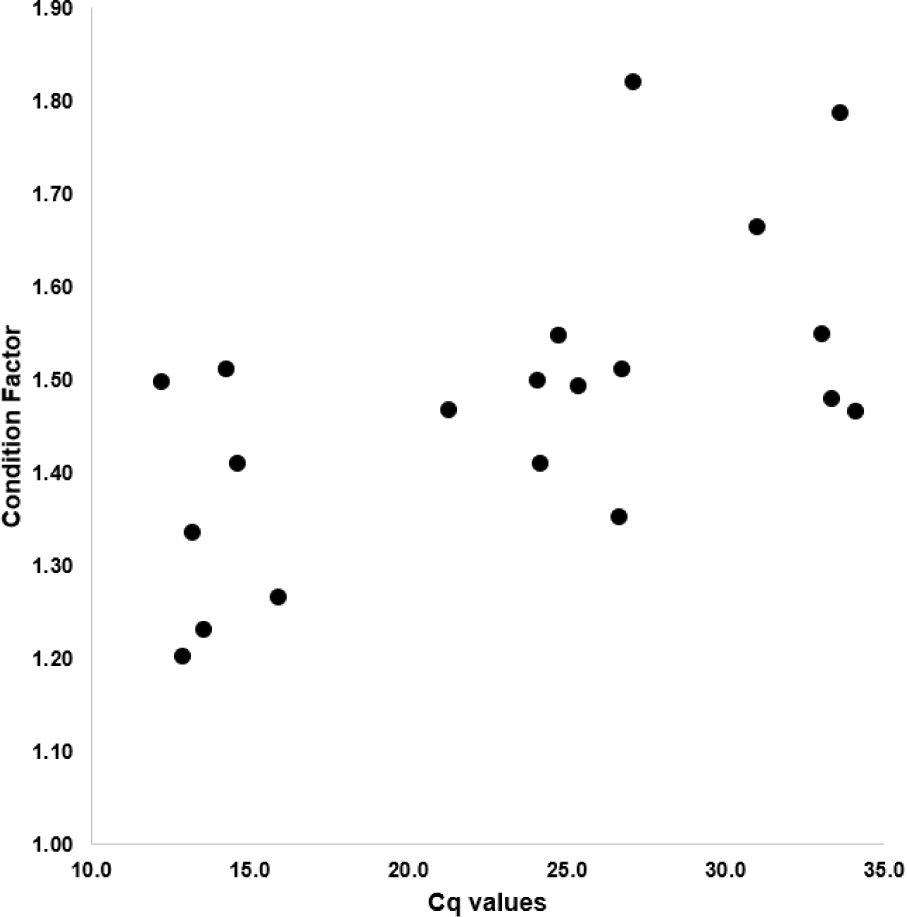
The CF of starry flounders investigated ranged from 1.20 to 1.82 and the cycle of quantification (Cq) value showed 12.2 to 34.1. As shown in Fig. 2 and Table 2, the CF was positively associated with Cq value (Fig. 2 and Table 2). Based on the Cq value, infection intensity was divided into three parts (high, moderate, low or negative). The mean CF in high infection intensity of parasites (Cq value: less than 23; more than 4.5 × 105 copies/100 mg) group is 1.36. The mean CF in moderate infection intensity of parasites (Cq value: 23–27; 4.5 × 105–2.1 × 104 copies/100 mg) group is 1.47, whereas the mean CF in low or negative (Cq value: more than 27; less than 2.1 × 104 copies/100 mg) group is 1.63 (Table 3).
Discussion
Recently, there has been a change in the aquaculture industry of Jeju island, Republic of Korea. The production of olive flounder decreased from 26,093 tons in 2016 to 21,463 tons in 2021, whereas the production of starry flounder increased from 446 to 2,610 tons between this period. The proportion of starry flounder production in 2021 accounted for approximately 10% of the total aquaculture production on Jeju island (KOSTAT, 2017; KOSTAT, 2022). This change can likely be attributed to the mass mortality of cultured olive flounder due to diseases (in particular, emaciation disease). Unfortunately, to date, there are no effective chemotherapeutics or vaccines against emaciation disease.
The mortality associated with emaciation disease of cultured olive flounder caused by E. leei shows seasonal changes in Korea. The mortality begins in September, and reaches its highest mortality rate from October to November, and decreases December onwards (Shin et al., 2018). The intensity of E. leei infection and the decrease in the relative CF in the olive flounder corresponded to these fluctuation (Sohn et al., 2021). In the present study, the detection rate of E. leei in cultured starry flounder also shows that of the similar fluctuation. Thus, we speculate that the onset of E. leei infection occurred at approximately the same time in cultured starry flounder and cultured olive flounder. However, Enteromyxum species require approximately two months to reach the developmental stage in the intestines after the initial onset of a natural infection (Sohn et al., 2021); hence, the infection of E. leei in cultured starry flounder may have likely begun in July.
Based on culturing experience, the members of fish farms in Jeju Island believed that the starry flounder are more resistant to emaciation disease than olive flounder. Although E. leei has been reported in more than 50 species of fish (Yanagida, 2017), there are no reports on starry flounder yet. Based on the results of the present study, the prevalence and intensity of E. leei in starry flounder were not lower than those in olive flounder that were infected naturally or experimentally. However, there was a clear difference in pathogenicity. The intensity of E. leei infection had a significant relationship with the CF of starry flounder, and the group with high infection intensity (Cq value: less than 23) of parasites had a significantly low CF compared to the group with low or negative (Cq value: more than 27). However, the difference in CF by the infection intensity of E. leei was a mean of more than 20%–30% in olive flounder (Shin et al., 2019, Shin et al., 2022; Sohn et al., 2021), whereas the mean was 16.6% in starry flounder. In addition, clinical signs of E. leei infection such as sunken eyes, fragile and semi-transparent intestine filled with mucous liquid, liver atrophy, and redness were not observed (data not shown). The mortality of cultured starry flounder (by E. leei) was also very low in the fish farm, and the feed intake did not decrease.
We suggest three possible explanations for the difference in pathogenicity 1) The difference of pathogenicity of E. leei depends on the host fish species. Previous studies have reported high pathogenicity in olive flounder, tiger puffer, sharpsnout sea bream Diplodus puntazzo, and low pathogenicity in gilt-head sea bream, red sea bream Pagrus major, and European sea bass Dicentrarchus labrax (Sitjà-Bobadilla & Palenzuela, 2012; Yanagida, 2017; Yanagida et al., 2008). The exact reason for the difference in pathogenicity depending on fish species has not been clarified; however, the difference in the development of E. leei depends on fish species or the genetic resistance of fish species (and strain) against myxosporean infection have been reported (Baerwald et al., 2011; Bartholomew, 1998; Sitjà-Bobadilla et al., 2007; Yanagida, 2017; Yanagida et al., 2004; Yasuda et al., 2005). 2) Co-infection with pathogenic bacteria and E. leei: some pathogenic bacterial infections (Genera Edwardsiella, Vibrio, and Streptococcus) from emaciated olive flounders have been reported (Choi et al., 2012; Kim et al., 2015; Yasuda et al., 2005), and the co-infection with Edwardsiella piscicida affected the pathogenicity of E. leei (Shin et al., 2022). No bacteria were isolated from the kidney and liver of starry flounder; however, previous studies have reported pathogenic bacteria (Photobacterium damselae subsp. piscicida and Streptococcus parauberis) in cultured starry flounder in Korea (Cho et al., 2008; Cho et al., 2013). Thus, further studies needed to reveal how pathogenic bacterial infections affect the pathogenicity of E. leei in starry flounders. 3) Acquired immunity: the starry flounders investigated in the present study were introduced to the fish farm in May 2021. Thus, the fish may have already been infected in the summer-autumn season of 2021 and acquired immunity. Fish that acquired immunity against E. leei infection were resistant to emaciation disease by E. leei (Picard-Sánchez et al., 2019; Yanagida et al., 2008).
Conclusion
The infection of Enteromyxum species was detected in cultured starry flounder and was identified as E. leei. Based on the statistical analyses, we revealed that high-intensity E. leei infection significantly decreased the CF of starry flounder. As the production of starry flounder and mortality due to emaciation disease in Jeju island increase, further studies (such as the factors that affect the pathogenicity of E. leei on starry flounder, host immunity, the vaccines, and chemotherapeutics against E. leei infection) are needed.
