Introduction
Ghana, a coastal country in West Africa bordered to the south by the eastern Atlantic Ocean, is notable for its rich marine biodiversity. According to the Convention on Biological Diversity (CBD) country profile, Ghana’s ecosystems exhibit high levels of species diversity and endemism, designating the country as a priority region for global biodiversity conservation (Caldecott et al., 1996). The country’s marine resources play a vital role in ensuring food security and driving socioeconomic development, with the fisheries sector forming a cornerstone of the national economy (Nunoo et al., 2014). This sector not only supports the livelihoods of millions of Ghanaians but also contributes significantly to the country’s GDP (Doku et al., 2018). Ghana’s marine and freshwater ecosystems host around 701 fish species, including approximately 499 in marine habitats and 58 associated with reef ecosystems (Froese & Pauly, 2024; accessed on November 5, 2024). Among these, 79 species are of commercial importance, including Chloroscombrus chrysurus (Atlantic bumper), Selene dorsalis (African moonfish), Pseudotolithus senegallus (cassava croaker), and Pagellus bellottii (red pandora), as documented in marine fish databases. These marine fish species are vital not only for ensuring domestic food security but also for supporting international trade, with certain species—particularly the cassava croaker— being exported to European and other global markets (Wehye et al., 2017). Hence, their substantial economic value highlights the necessity for effective management strategies grounded in integrated scientific approaches and principles of sustainable exploitation (Takyi et al., 2023).
Beyond traditional morphology-based methods, DNA barcoding has emerged as a reliable and crucial tool for genetic monitoring of fish species across diverse marine ecosystems (Afriyie et al., 2019). This technique targets a 650-base-pair region of the mitochondrial cytochrome c oxidase subunit I (COI) gene, which serves as the standard marker for species differentiation (Kundu et al., 2019). DNA barcoding not only aids in resolving taxonomic challenges and revealing cryptic species diversity but also addresses common misidentification issues due to sexual polymorphism and morphological plasticity (van Ginneken et al., 2017). Moreover, because many fish undergo processing before reaching commercial markets, traditional taxonomic identification can be challenging. DNA barcoding has proven effective in detecting mislabeling and insufficient labeling of commercially significant marine species (Agyeman et al., 2021). The genetic data obtained from barcoding further contribute to fish stock assessments, improvements in aquaculture, and conservation efforts for globally threatened species (Ferrette et al., 2019). Additionally, DNA barcoding aids in assessing population genetic diversity, providing valuable insights for species conservation and management (Kundu et al., 2023).
Recent research on commercially significant fish species in Ghana has focused on important areas such as species-specific growth and mortality rates, reproductive patterns, population dynamics, socio-economic impacts of artisanal fisheries, and habitat distribution (Aggrey-Fynn & Sackey-Mensah, 2012; Amponsah et al., 2023). Additional studies have highlighted the challenges faced by small-scale fisheries, including overexploitation, habitat degradation, and illegal, unreported, and unregulated fishing, which threaten both fish stock sustainability and the economic resilience of local communities (Ameyaw et al., 2021; Okafor-Yarwood, 2019). Much of this research has relied on morphological and morphometric data, which leaves a gap in understanding the genetic diversity and finer-scale population dynamics of key commercial species native to Ghana. Some genetic studies, however, have documented native fish diversity and assessed the presence of invasive species in the region (Afriyie et al., 2019; Elsaied et al., 2021). Initiatives such as the FISH-BOL project have also contributed to the development of a comprehensive database for African fish species, supporting broader genetic monitoring and conservation efforts (Elsaied et al., 2021; Swartz et al., 2008).
Despite recent advancements, there remains a substantial gap in molecular data for indigenous Ghanaian fish species—data critical for accurate biodiversity assessment, aquaculture development, and sustainable conservation efforts. To address this research gap, the present study aims to: (i) collect key commercially important fish species from Ghana between 2023 and 2024; (ii) generate molecular data to enable rapid and accurate species-level identification; and (iii) estimate genetic distances, perform phylogenetic analyses with species delimitation, and assess population genetic structure. The resulting genetic divergence, phylogenetic relationships, estimated operational taxonomic units (OTUs), haplotypic university as well as networks will together provide foundational phylogeographic insights into these key species. Furthermore, the generated dataset will contribute to the development of a robust molecular reference library for Ghanaian fish species, thereby supporting future ichthyological research, biodiversity monitoring, and conservation efforts in the region.
Materials and Methods
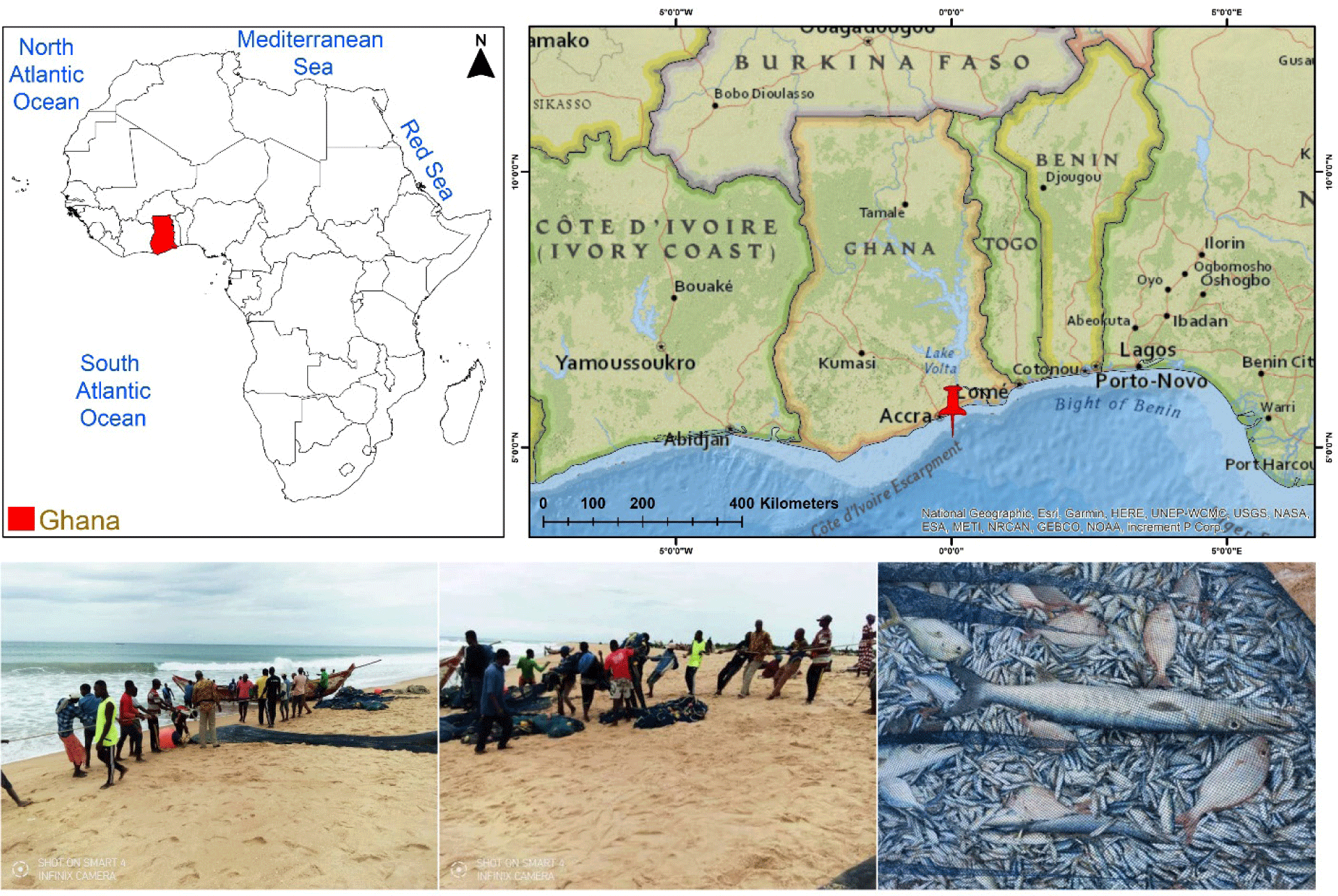
A total of ten fish samples were collected from the Atlantic Ocean at Tema Fishing Harbour, located along Ghana’s coast at coordinates 5.645007°N, 0.016947°E (Fig. 1). Species identification and taxonomic classification were verified using morphological keys documented in prior taxonomic studies, as detailed in Eschmeyer’s Catalog of Fishes (Fricke et al., 2024). Muscle tissue samples were extracted from each specimen and preserved in 70% ethanol, while voucher specimens were fixed in 10% formaldehyde with unique identification numbers assigned at the Fisheries Scientific Survey Division of the Fisheries Commission, Ghana. To prevent DNA degradation and minimize microbial contamination, tissue samples were sealed in centrifuge tubes with parafilm and transported in a temperature-controlled icebox. Upon arrival, samples were transferred to the Molecular Physiology Laboratory at Pukyong National University, Korea, for subsequent molecular analysis. All experimental procedures were approved by the host institute Pukyong National University (Approval No. PKNUIACUC-2025-16) and conducted in accordance with the ARRIVE 2.0 guidelines (https://arriveguidelines.org; du Sert et al., 2020).
Genomic DNA was extracted using the AccuPrep® DNA Extraction Kit (Bioneer, Daejeon, Korea), following standard protocols. The quality and concentration of extracted DNA were measured with a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). In brief, 30 mg of tissue was homogenized in 600 μL of 1× lysis buffer using a TissueLyser II (Qiagen, Hilden, Germany) for 60 seconds. To disrupt cellular membranes and degrade proteins, 100 μL of sodium dodecyl sulfate and 20 μL of proteinase K were added, followed by overnight incubation at 60°C. After incubation, 500 μL of GC buffer and 300 μL of isopropanol were added to precipitate the DNA. The mixture was transferred to a spin column and centrifuged at 8,000×g for one minute. Remaining biomolecules were removed with wash buffers 1 and 2, and DNA was eluted in 50 μL of TE buffer. The mitochondrial COI gene was amplified using the primer pair FISH-BCH (5¹-TAAACTTCAGGGTGACCAAAAAATCA) and FISH-BCL (5¹-TCAACYAATCAYAAAGATATYGGCAC; Baldwin et al., 2009). Polymerase chain reaction (PCR) was conducted in a 30 μL reaction mix on a Takara thermal cycler, containing 1 μL each of forward and reverse primers, 0.9 μL of 3% dimethyl sulfoxide, 19.9 μL of sterilized deionized water, 3 μL of 10 × ExTaq Buffer, 0.2 μL of Ex Taq HS enzyme, 3 μL of dNTPs, and 1 μL of a 1/10 diluted DNA template. The thermal profile included an initial denaturation at 94°C for 3 minutes, followed by 40 cycles of denaturation at 94°C for 30 seconds, annealing at 50°C for 30 seconds, extension at 72°C for 1 minute, and a final extension at 72°C for 5 minutes. PCR products were purified with the AccuPrep® PCR/Gel Purification Kit (Bioneer) and sequenced bidirectionally using a 96-capillary ABI PRISM 3730XL Analyzer at Macrogen (Daejeon, Korea). Noisy segments were trimmed from chromatograms using SeqScanner v1.0 (Applied Biosystems, Waltham, MA, USA). The COI sequences were edited and aligned with ClustalX in MEGA X (Kumar et al., 2018; Thompson et al., 1997), then verified through nucleotide BLAST (https://blast.ncbi.nlm.nih.gov) against GenBank for species identification. Final sequences were submitted to GenBank to obtain unique accession numbers.
For genetic analysis, a total of 138 sequences representing target species were retrieved from the global GenBank database (Table S1). This dataset included 10 newly sequenced DNA barcodes from Ghanaian fish species, along with a single sequence of Apristurus ampliceps (Accession No. EU398546) used as an outgroup taxon. Geographic information associated with each sequence was also gathered to aid demographic interpretation. MEGA X was used to estimate variable and conserved nucleotide counts within the dataset. Genetic distances within and between species were calculated using the Kimura 2-parameter (K2P) model. Haplotype diversity (Hd) was assessed for each species by aligning both newly generated and GenBank sequences, creating separate datasets for each species studied. Estimates of haplotype count, Hd, and nucleotide diversity (π) were computed using DnaSP v6 (Rozas et al., 2017), and haplotype networks were constructed with TCS in PopArt (Leigh & Bryant, 2015). The best-fit model was determined to be ‘GTR+G+I’ based on the lowest Bayesian Information Criterion (BIC) score, identified through the use of PartitionFinder 2 on the CIPRES Science Gateway v3.3, and JModelTest v2 (Darriba et al., 2012; Lanfear et al., 2016; Miller et al., 2015). A Bayesian topology was built using Mr. Bayes 3.1.2, with the model parameter nst = 6, and Metropolis-coupled Markov Chain Monte Carlo (MCMC) chains configured with one cold and three hot chains. The analysis was run for 10,000,000 generations, with trees sampled every 100th generation, and the first 25% of the samples discarded as burn-in (Ronquist & Huelsenbeck, 2003). The BA phylogeny was visualized using iTOL (https://itol.embl.de/) to enhance clarity and presentation (Letunic & Bork, 2007). Additionally, the Assemble Species by Automatic Partitioning (ASAP) approach was employed for species delimitation, well-suited for single-locus sequence dataset (Puillandre et al., 2021).
Results and Discussion
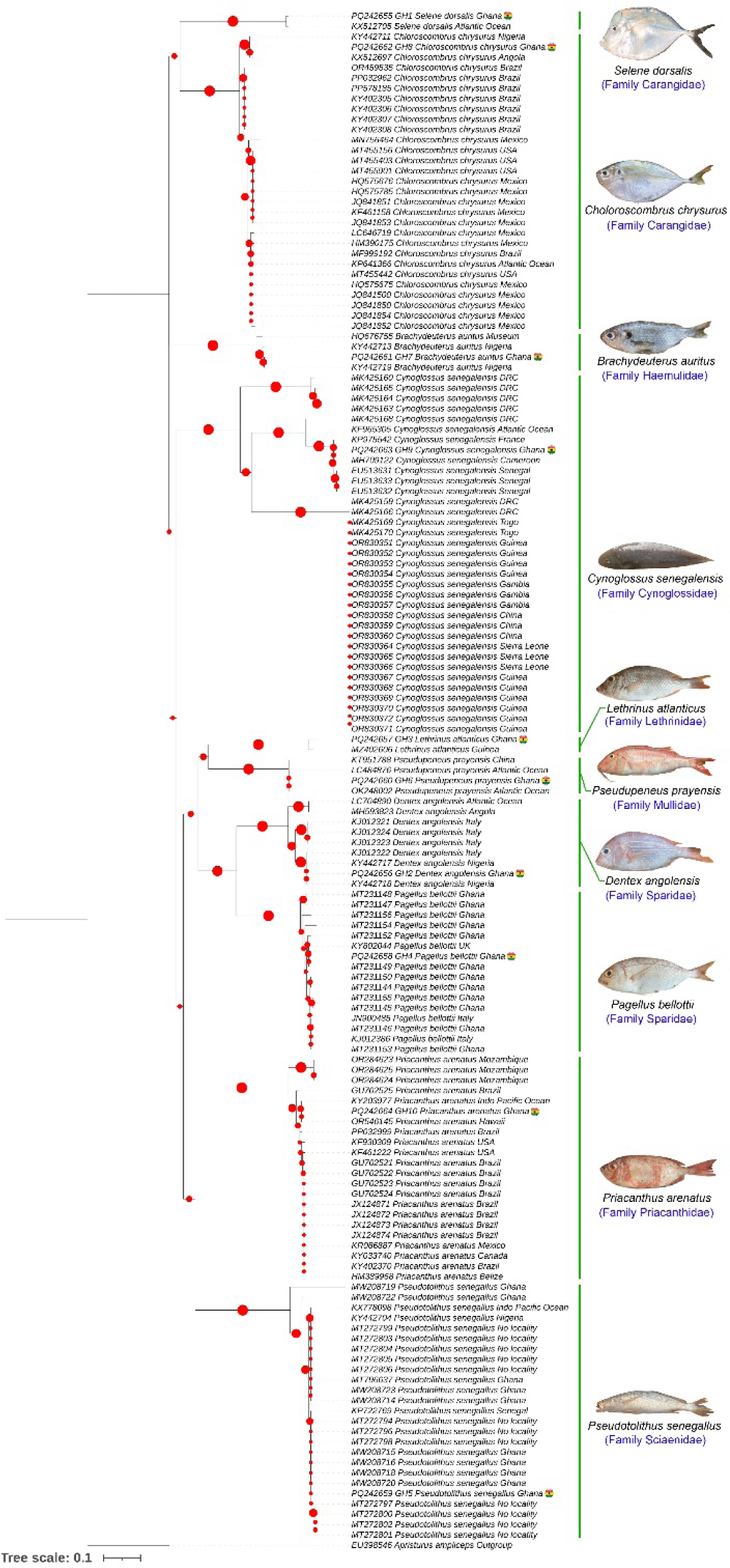
Morphological characteristics identified all the fish specimens in to 10 species collected from the Eastern Atlantic Ocean, Ghana. Sample ‘GH1’ was identified as S. dorsalis and ‘GH8’ as C. chrysurus, both classified in the family Carangidae, subfamily Caranginae. The African moonfish, S. dorsalis is distributed from the Southwestern Mediterranean to the Eastern Atlantic, while C. chrysurus has a broader distribution from the Western to the Eastern Atlantic. The ‘GH2’ and ‘GH4’ samples were identified as Dentex angolensis and P. bellottii, respectively, both in the family Sparidae. D. angolensis is endemic to the Eastern Atlantic, while P. bellottii occurs in both the Mediterranean Sea and Eastern Atlantic Ocean. Sample ‘GH3’ was identified as Lethrinus atlanticus, classified under the family Lethrinidae and subfamily Lethrininae, with distribution in the Eastern Atlantic. The ‘GH5’ specimen was identified as P. senegallus of the family Sciaenidae, distributed in the Eastern Atlantic. The ‘GH6’ sample was identified as Pseudupeneus prayensis, family Mullidae, found in both the Mediterranean and Eastern Atlantic, whereas the ‘GH7’ Sample was identified as Brachydeuterus auritus in the family Haemulidae, distributed in the Eastern Atlantic. The sample ‘GH9’ was identified as Cynoglossus senegalensis, family Cynoglossidae, also found in the Eastern Atlantic, while the sample ‘GH10’ was identified as Priacanthus arenatus, family Priacanthidae, which has a wide distribution from the Western to the Eastern Atlantic.
The DNA barcode sequences generated for the 10 species were deposited in GenBank under accession numbers PQ242655−PQ242664. The sequence for S. dorsalis (GH1: PQ242655) matched the corresponding GenBank entry (KX512705) with 99.25% similarity (Damerau et al., 2018). Similarly, D. angolensis (GH2: PQ242656) showed 99.68% similarity with the database sequence (KJ012322; Armani et al., 2015), and L. atlanticus (GH3: PQ242657) was 98.60% similar with the database sequence (MZ402606; Allen et al., 2021). The generated DNA barcode sequence of P. bellottii (GH4: PQ242658) aligned with the database sequence (KJ012386) at 99.69% similarity (Armani et al., 2015), while P. senegallus (GH5: PQ242659) showed 99.53% similarity with the unpublished database sequence (MT796637). The sequence for P. prayensis (GH6: PQ242660) demonstrated 99.84% similarity with the database sequence (LC484876; Li et al., 2020), and B. auritus (GH7: PQ242661) had 98.76% similarity with the database sequence (HQ676755; Sanciangco et al., 2011). The DNA barcode sequence of C. chrysurus (GH8: PQ242662) showed complete alignment (100%) with the database sequence (KY442711; Bolaji et al., 2023). For C. senegalensis (GH9: PQ242663), a 99.85% similarity was observed with the database sequence (MH709122; Gietbong et al., 2018), and P. arenatus(GH10: PQ242664) aligned at 99.70% similarity with the database sequence (OR546145; Hoban et al., 2022).
The analyzed COI gene dataset (652 bp) for the 10 marine fish species revealed an overall mean genetic distance of 25.6%. Most species exhibited < 2% mean intra-species genetic distance, except for C. senegalensis (9.93%) and D. angolensis (2.56%; Table 1). Mean inter-species genetic distances ranged from 16.55% between D. angolensis and P. bellottii to 35.29% between P. bellottii and C. senegalensis (Table 1). The lower inter-species distance observed between D. angolensis and P. bellottii is likely due to their shared lineage within the family Sparidae. Similarly, two species in the family Carangidae, C. chrysurus and S. dorsalis, showed a mean inter-species genetic distance of 20.37%. The NJ topology distinctly grouped each species into their respective families, demonstrating monophyletic clustering of both the newly generated sequences and reference sequences retrieved from GenBank (Fig. 2). Additionally, the ASAP species delimitation approach successfully partitioned all studied species, identifying OTUs within C. senegalensis (Fig. 3). Of the 10 species studied, most showed more than one haplotype in the COI gene, with the exception of P. prayensis. The limited genetic diversity detected in P. prayensis COI sequences, derived from geographically distant sites such as China and unidentified locations in the Atlantic Ocean, indicates a potential influence of human-mediated translocation, likely facilitated by international fisheries trade. The highest Hd was observed in Brachydeuterus auratus, L. atlanticus, and S. dorsalis. P. bellottii exhibited the greatest number of haplotypes, with a Hd of 0.9500 and nucleotide diversity (π) of 0.2488 (Table 2).

In the haplotype network analysis, the DNA barcode of C. chrysurus generated from Ghana shared a haplotype with the COI sequence from Nigeria, both countries located along the northeastern Atlantic coast. In contrast, C. chrysurus from Angola, situated in the southeastern Atlantic, differed by a single variable nucleotide from the Ghanaian and Nigerian haplotypes. This minimal genetic divergence may be influenced by ocean currents and larval dispersal patterns facilitating gene flow between the neighboring West African countries. Additionally, barcodes from C. chrysurus populations in Brazil, North America, and Mexico showed distinct haplotypes compared to those from Africa (Fig. 4A). The Ghanaian sequence of C. chrysurus maintained a genetic distance of 3.9% from other GenBank sequences sourced widely across the western Atlantic. For P. bellottii, the barcode from Ghana shared a haplotype with sequences from Italy and Liverpool, UK, which may be influenced by the human-mediated transport through fisheries trade. Notably, multiple haplotypes were observed within the Ghanaian sequences of P. bellottii, indicating regional genetic differentiation (1.7%) or possible sample origins from distant marine environments (Fig. 4B). Interestingly, the barcode of C. senegalensis, a species broadly distributed across the southwestern Mediterranean and eastern Atlantic, shared a haplotype with sequences from France and Cameroon. Meanwhile, sequences from the Democratic Congo showed distinct haplotypes and high nucleotide variability from those of Ghana. Other African regions, including Gambia, Guinea, Togo, Senegal, and Sierra Leone, also presented unique haplotypes for C. senegalensis (Fig. 4C). The unusually high intra-species genetic distance (21.1%) observed in C. senegalensis may reflect potential misidentifications within the database, warranting further investigation. The Ghanaian barcode of D. angolensis also showed a unique haplotype, closely associated with Nigerian sequences, yet exhibited significant nucleotide variation (4.2% genetic distance) when compared to sequences from Italy, Angola, and other Atlantic regions. A sequence from Italy closely aligned with the Ghanaian haplotype, which may reflect sampling from international markets with undocumented source origins (Fig. 4D).
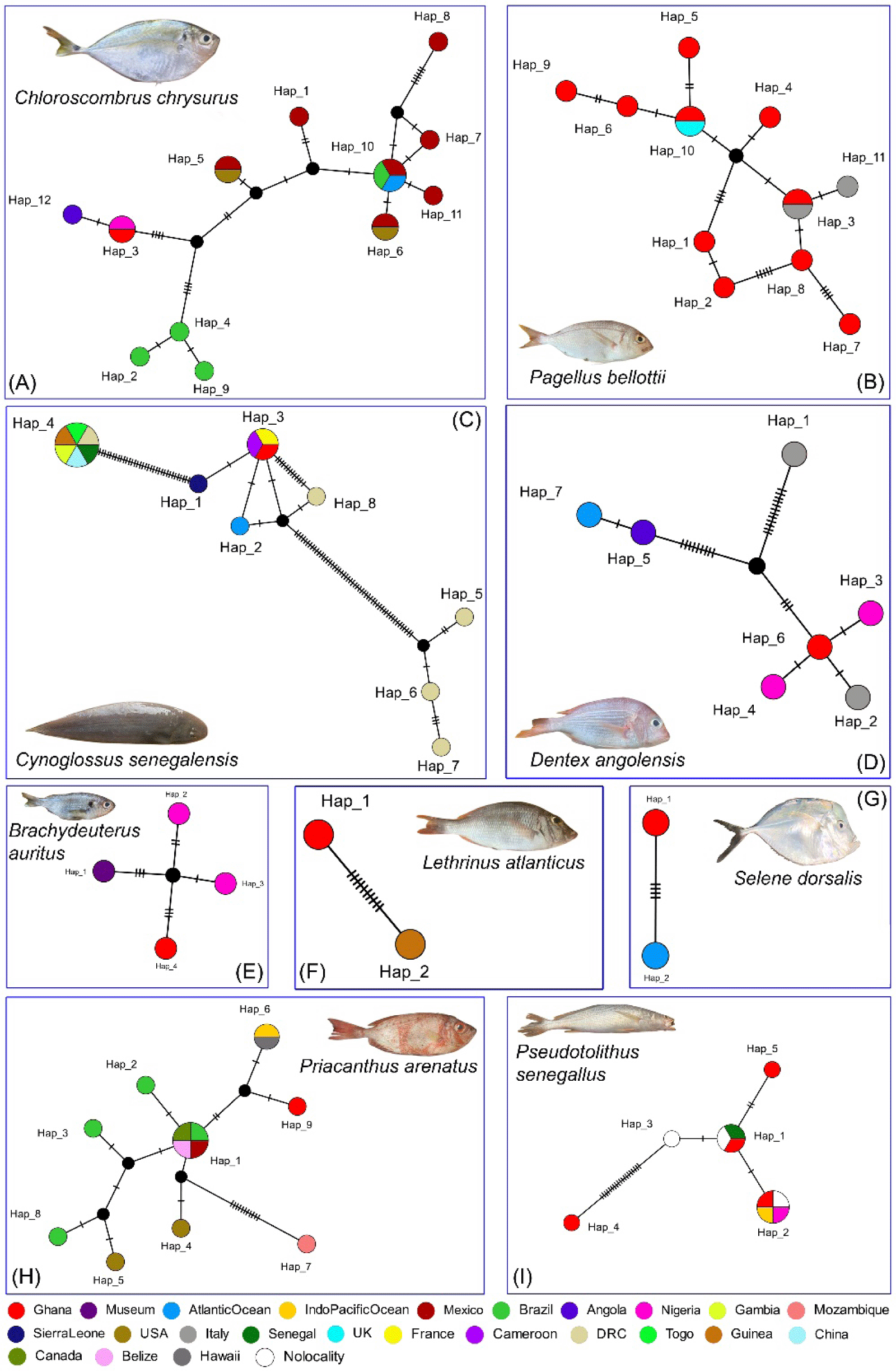
DNA barcode data for three commercially important fish species (B. auratus, L. atlanticus, and S. dorsalis) remain scarce in the global GenBank database. Although this study generated COI barcodes for these species from Ghana, the current population genetic structures are incomplete due to their wide distribution in the eastern Atlantic and the limited availability of genetic information. Comparisons with the available sequences revealed genetic distances of 1.1% for B. auratus (Nigeria), 1.6% for L. atlanticus (Guinea), and 0.8% for S. dorsalis (Atlantic Ocean), providing insight into the distinct Ghanaian haplotypes of these species (Fig. 4E–4G). For P. arenatus, the Ghanaian barcode showed a unique haplotype, closely related to a shared haplotype from the Indo-Pacific Ocean and Hawaii. Sequences from Brazil, Canada, Belize, Mexico, the USA, and Mozambique presented distinct haplotypes, with a maximum genetic distance of 3.8% (Fig. 4H). P. senegallus from Ghana also displayed a unique haplotype network, comprising two distinct and two shared haplotypes, with sequences from Senegal, the Indo-Pacific Ocean, and Nigeria, showing a genetic distance of 8.6%.
Overall, the haplotype networks reveal a distinct genetic composition and population structure in the commercially important fish species studied from Ghana, compared to those from other African countries in the eastern Atlantic and European regions in the Mediterranean Sea. This observed Hd may result from the existence of different populations inhabiting reef-associated ecosystems in the Atlantic Ocean. Additionally, as the Ghanaian fish samples were primarily collected from a fishing harbor, it is possible that some were sourced from distant waters, which may explain the observed nucleotide variation or similarities in shared haplotypes across distant locations (Fig. 4I). Notably, the high intra-species genetic distances and multiple haplotypes identified in five species—C. chrysurus, C. senegalensis, D. angolensis, P. arenatus, and P. senegallus—suggest the potential presence of cryptic diversity within these populations.
The current genetic analysis using DNA barcoding presents a robust and reliable method for assessing the genetic diversity and population structure of commercially important fish species from Ghana. This molecular approach has become indispensable for exploring genetic variability in marine biodiversity, as it enables accurate species identification and provides critical insights into the genetic composition of conspecific populations across open marine environments, thereby offering essential information for effective conservation and management efforts (Krishnamurthy & Francis, 2012). Given the limitations of morphology-based species identification—particularly in cases of phenotypic similarity and cryptic speciation—this study underscores the significance of molecular tools in establishing a comprehensive barcode reference library for Ghanaian fish species, thereby facilitating more precise species delineation (Elsaied et al., 2021; Kundu et al., 2019; van Ginneken et al., 2017). The observed intra- and interspecific genetic distances among the studied species suggest potential signals of speciation and population substructure, possibly driven by environmental gradients or geographic isolation. Conversely, the detection of low genetic diversity in certain species may reflect recent population bottlenecks or limited gene flow, as previously reported (Paz-Vinas et al., 2015). Therefore, more extensive and region-wide studies are warranted to comprehensively investigate the population structure of these marine species in the Atlantic Ocean, particularly to elucidate patterns of population connectivity. High levels of genetic connectivity are indicative of healthy gene flow, which is instrumental for maintaining genetic diversity. In contrast, isolated populations are more susceptible to inbreeding depression and may exhibit reduced resilience to environmental stressors (Neff et al., 2011; Reisenbichler et al., 2003). Beyond conservation implications, the genetic data generated in this study may also contribute to the development of sustainable aquaculture practices in Ghana by enabling the management of genetically distinct populations and informing targeted breeding programs. Overall, DNA barcoding-based assessments of genetic diversity can reveal distinct genetic clusters within Atlantic marine fish populations, identify biodiversity hotspots or unique evolutionary lineages, and support the formulation of targeted conservation and resource management strategies for Ghana’s marine ecosystems and the broader Atlantic region.
Limitations and Recommendation
This preliminary genetic analysis of commercially important fish species from Ghana, conducted over a short duration of approximately 1.5 years, is based on a single representative specimen per species from the study location. Initial morphological assessments were conducted based on literature reviews and key diagnostic characteristics. The primary objectives of the study—species identification, estimation of genetic distances, and phylogenetic analysis—were addressed using the limited number of sequences obtained from the sampled individuals. However, to enhance the robustness of these analyses, additional reference sequences for the corresponding species were retrieved from the GenBank database. Except for S. dorsalis and L. atlanticus, most of the other target species had four or more sequences in the database, allowing for reasonably reliable haplotypic analyses. Based on the findings from this pilot investigation, the study highlights the need for more extensive sampling of marine fish species from Ghana. This should include detailed morphological assessments of individuals from multiple populations, coupled with the generation of both mitochondrial and nuclear genetic markers as well as genome-wide data, to more accurately resolve the population genetic structure and detection of cryptic diversity of these species in the Atlantic Ocean. Furthermore, this study employed the ASAP species delimitation method, which is suitable for single-locus sequence datasets. However, future studies incorporating a broader geographic range and multiple sequences per species should consider applying a combination of species delimitation methods, such as Barcode Index Numbers (BINs) from the Barcode of Life Data System (BOLD), Automatic Barcode Gap Discovery (ABGD), and coalescent-based approaches like General Mixed Yule Coalescent (GMYC) and Poisson Tree Processes (PTP), using both Bayesian and Maximum-likelihood frameworks. In addition to expanding genetic analyses, the integration of oceanographic data—including current patterns, larval dispersal routes, regional connectivity, and human-mediated transport—will be critical for developing a comprehensive phylogeographic understanding of the target fish species within the open environment in Atlantic Ocean.
Conclusion
This study underscores the importance of DNA barcoding, haplotype analysis, and phylogenetic approaches in elucidating the genetic diversity and evolutionary relationships of commercially significant fish species from Ghana. The genetic data successfully identified and differentiated the target species, revealing unique Hd within Ghana and uncovering cryptic diversity in several species. By integrating this genetic information, Ghana can promote sustainable aquaculture practices through the identification of distinct wild populations from open marine environment, which would be beneficial for mitigating inbreeding risks and enhancing species conservation strategies by preserving the genetic integrity. Collectively, these findings lay a solid foundation for the sustainable management of fisheries and the conservation of biodiversity in Ghana’s marine ecosystems. Furthermore, the generated genetic data serve as a valuable reference for future research, supporting the development of a national barcode reference library for Ghana to enhance species identification and biodiversity assessments.
