Introduction
Innate immunity is an important defense system in fish species, dealing with a broad spectrum of pathogens. Lysozymes are well-known players in the innate immune mechanisms of fish, performing diverse functional roles, including antimicrobial, immunomodulatory, anti-inflammatory, and antitumor activities in living organisms (Kokoshis et al., 1978; Lee-Huang et al., 1999; Nilojan et al., 2017; Sava et al., 1988). They are distributed in a wide range of tissues and body fluids, such as mucus, lymph, and plasma of both freshwater and marine fish (Saurabh & Sahoo, 2008). Lysozyme activity and expression are reportedly modulated under different abiotic (degree of stress, pH, water temperature, content of pollutants, and environmental salinity) and biotic (sexual maturity stages, sex, age, and size) conditions (Saurabh & Sahoo, 2008). Lysozymes protect organisms from microbial infections by destroying the bacterial cell wall by hydrolyzing the β-1, 4-glycosidic bond in the peptidoglycan layer (Umasuthan et al., 2013); therefore, lysozymes exert a powerful antibacterial function against gram-positive bacteria by solubilizing the peptidoglycan layer. However, gram-negative bacteria are resistant to cationic antimicrobial peptides, as their peptidoglycan layer is less vulnerable to hydrolysis by lysozymes (Hancock & Scott, 2000).
There are three types of lysozymes: a-type (animal), p-type (plant), and m-type (microbial) (Li et al., 2018). Depending on the amino acid sequence and their chemical and biological properties, animal-type lysozymes are further divided into three subclasses: c-type (chicken), g-type (goose), and i-type (invertebrate) (Pridgeon et al., 2013). The c- and g-type lysozymes were discovered in both invertebrates and vertebrates, whereas i-type lysozymes were identified only in invertebrates. Generally, the secondary structure of lysozymes comprises four alpha helices and five beta strands (Prilusky et al., 2011). All three types of lysozymes share a similar tertiary structure but have low homogeneity in the underlying structure (Li et al., 2019). The c-type lysozyme was first extracted from chicken egg white, and the g-type lysozyme was identified in egg white from Embden goose (Xie et al., 2019). Furthermore, c- and g-type lysozymes have been previously reported in various marine animals, such as Yangtze sturgeon (Acipenser dabryanus) (Zhang et al., 2018), longtooth grouper (Epinephelus bruneus) (Harikrishnan et al., 2011), orange-spotted grouper (Epinephelus coioides) (Wei et al., 2012), Manila clam (Ruditapes philippinarum) (Wei et al., 2018), Japanese flounder (Paralichthys olivaceus) (Hikima et al., 2001), seahorse (Hippocampus abdominalis) (Ko et al., 2016), and turbot (Scophthalmus maximus) (Zhao et al., 2011). However, very few reports reveal the in vitro antibacterial activity of c-type lysozyme and g-type-like lysozyme in ornamental fish (Smith et al., 2012).
As the demand for ornamental aquaculture grows and the aquarium trade increases, many studies have focused on the growth and immune systems of ornamental fish to enable more systemic aquacultural production (Avella et al., 2007). Yellowtail clownfish (Amphiprion clarkii) is one of the most frequently demanded members of Amphiprioninae in aquarium trade markets. It is found in coral reefs or shallow lagoons in the Indian Ocean, Pacific Ocean, and the Red Sea, where the water temperature is approximately 22°C–29°C (Yanagisawa & Ochi, 1986). Moreover, it inhabits the ocean around Jeju Island in South Korea. Nearly 90% of all marine ornamental species are captured by fishing. To develop sustainable ornamental fish aquaculture, it is essential to implement a proper breeding system and reduce lethal infections such as vibriosis, edwardsiellosis, and lymphocystis. Moreover, clownfish are now emerging as potential model organisms of interest for developmental biology, ecology, and evolutionary science (Roux et al., 2020). Therefore, understanding the innate immune mechanisms in A. clarkii can help develop useful strategies for preventing diseases in ornamental fish aquaculture. In this study, two lysozymes, c-type lysozyme (AcLysC) and g-like-type lysozyme (AcLysG-like) were identified and characterized at the molecular level to elucidate their functional roles in A. clarkii.
Materials and Methods
Healthy A. clarkii with an average body weight of 20 g and an average body length of 10 cm were obtained from the Choryang Aquarium, Busan, Korea. The fish were acclimated in laboratory aquarium tanks (300 L) at a constant temperature of 28°C and 32 PSU (practical salinity unit) for one week prior to experiments. During acclimatization, fish were fed a commercial fish feed twice daily and deprived of feed two days prior to the experiment.
Tissues were collected from A. clarkii (n = 5) to perform the transcriptome and tissue-specific expression analyses of AcLysC and AcLysG-like. First, conventional anesthetics (MS-222; 40 mg/L) were used to anesthetize healthy A. clarkii. Then, the fish were dissected, and the head kidney, intestine, heart, liver, brain, gill, spleen, kidney, muscle, skin, and stomach tissues were collected. Blood was also sampled using sterile syringes coated with heparin sodium salt (Sigma-Aldrich, St. Louis, MO, USA). Peripheral blood cells were immediately harvested by centrifugation at 3,000×g for 10 min at 4°C. All tissues were immediately snap-frozen in liquid nitrogen and stored at −80°C until RNA isolation.
To evaluate the modulation of AcLysC and AcLysG-like against immune stimulation, two chemical stimulants and a live bacterial pathogen were used in time-course experiments. Briefly, the acclimatized A. clarkii were randomly divided into four groups (each group including four animals). Fish of three groups constituting the test groups for this experiment were administered a single intraperitoneal injection of either lipopolysaccharide (LPS; 2.5 μg/g, Sigma-Aldrich), polyinosinic: polycytidylic acid (Poly I:C; 2.5 μg/g, Sigma-Aldrich), or Vibrio harveyi (1 × 103 CFU/μL) suspended in 100 μL of phosphate-buffered saline (PBS). Fish in the control group were injected with 100 μL of PBS. Tissues were collected at 0, 6, 12, 24, 48, and 72 h post-injection (p.i.). All tissues were snap-frozen in liquid nitrogen and stored at −80°C until RNA isolation. All experimental procedures involved in this study were reviewed and approved by the Animal Care and Use Committee of the Jeju National University.
To prepare the transcriptome library, total RNA was extracted from collected tissues. RNA extraction was performed using TRIzol (Invitrogen, Waltham, MA, USA), followed by clean-up with a RNeasy spin column (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The extracted RNA was assessed for quality, quantified using a 2100 Expert Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), and sequenced at Insilicogen (Yongin, Korea). The transcriptome database of A. clarkii was constructed using the Illumina NovaSeq6000 and assembled de novo at Insilicogen.
For expression analysis (tissue distribution, n = 5; immune challenge, n = 4), total RNA was extracted, and the purity of the isolated RNA was examined with 1.5% agarose gel electrophoresis. The concentration was determined based on the absorbance at 260 nm and 280 nm, measured using a µDrop Plate (Thermo Scientific, Waltham, MA, USA). First-strand cDNA was synthesized in a 20 μL reaction mixture containing a pool of RNA (2.5 μg of RNA from tissue distribution samples, 2.0 μg of RNA from immune challenge samples) with PrimeScript™ II 1st strand cDNA Synthesis Kit (TaKaRa, Shiga, Japan) according to the recommended procedures. The synthesized cDNA was diluted 40-fold in nuclease-free water and stored in a freezer at −80°C until further use.
The cDNA sequences of AcLysC (MK835689) and AcLysG-like (MW199185) were identified from the established A. clarkii transcriptomic database using the NCBI BLAST tool based on the sequence alignments with respective lysozyme homologs in NCBI-GenBank database. The open reading frame sequences (ORFs) of AcLysC and AcLysG-like were obtained using the NCBI ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/). Then, the corresponding amino acid sequences were verified using the NCBI BLAST program (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Signal peptides of AcLysC and AcLysG-like were analyzed using the SignalP 5.0 online tool (http://www.cbs.dtu.dk/services/SignalP/). The domain organization and signature motifs in the AcLysC and AcLysG-like amino acid sequences were identified using the NCBI conserved domain database (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and Motif Scan (https://myhits.sib.swiss/cgi-bin/motif_scan) online server. Moreover, multiple sequence alignments of deduced AcLysC and AcLysG-like protein sequences with corresponding homologs were performed using the CLC Main Workbench (version 8.0) software. A phylogenetic tree comprising different homologous sequences of AcLysC and AcLysG-like was constructed using the neighbor-joining method embedded in the MEGA 6.0 software. The reliance value for the phylogenetic tree was obtained by replicating bootstrap trials 5,000 times.
Quantitative real-time polymerase chain reaction (qPCR) was carried out using a Thermal Cycler DiceTM TP 950 (TaKaRa) in a 10 μL reaction volume containing 3 μL of diluted cDNA template, 5 μL of 2 × Ex TaqTM SYBR premix (TaKaRa), 0.4 μL each of the forward and reverse primers (10 pmol/μL), and 1.2 μL of PCR-grade H2O (Table 1). The qPCR thermal cycles included one cycle of 95°C for 10 s, followed by 45 cycles of 95°C for 5 s, 58°C for 10 s, 72°C for 20 s, and a final single cycle of 95°C for 15 s, 60°C for 30 s, and 95°C for 15 s. Each assay was conducted in triplicates. The 2−ΔΔCt method was used to calculate the relative expression of each gene (Livak & Schmittgen, 2001). Elongation factor 1-beta (Ef1β) of A. clarkii (MN970208.1) was used as the internal control gene for all qPCR assays. Furthermore, mRNA expression levels of the immune-stimulated group were normalized to those of relevant PBS-injected control at each time point. The obtained results were represented as mean ±SD. Expression level at 0 h (uninjected control) was used as the basal level reference for the expression profiles of immune-stimulated fish. An independent, two-tailed t-test was carried out to identify the statistically significant fold expression values of immune-stimulated animals at p < 0.05 and p < 0.01.
Primer pairs were designed for PCR amplification of the ORFs of AcLysC and AcLysG-like with suitable restriction sites (Table 1) for cloning into the expression vector pMAL-c5X (NEB, Ipswich, MA, USA). The coding sequence was amplified from cDNA using Ex Taq polymerase (TaKaRa). Amplified PCR products and the pMAL-c5X vector were separately digested with respective restriction enzymes. Digested ORFs were separately ligated into the pMAL-c5X vector using DNA Ligation Mighty Mix (TaKaRa). The recombinant expression vector for each gene was transformed into Escherichia coli DH5α cells. Successful transformants were identified by colony PCR and confirmed by sequencing (Macrogen, Seoul, Korea). Sequence-confirmed colonies were subjected to plasmid extraction using the AccuPrep® Plasmid MiniPrep DNA Extraction Kit as per the vendor’s protocol (Bioneer, Daejeon, Korea).
The recombinant AcLysC and AcLysG-like plasmids were transformed into E. coli ER2523 cells. Recombinant bacterial cells were cultured in 500 mL Luria-Bertani broth supplemented with 2.0 g of glucose and 100 μg/mL ampicillin. Cells were grown at 37°C in a shaking incubator (200 rpm) until the optical density (OD) at 600 nm reached 0.7. At the optimal OD, protein expression was induced by adding a final concentration of 0.25 mM isopropyl-b-thiogalactopyranoside (IPTG) and incubating for 8 h at 25°C. Bacterial cultures were then cooled on ice for 30 min. Subsequently, bacteria were harvested by centrifugation at 4,000×g for 30 min at 4°C, and the obtained cell pellets were resuspended in 25 mL of column buffer (20 mM Tris-HCl, pH 7.4, 200 mM NaCl, 0.5 M EDTA). The fusion protein with maltose-binding protein (MBP) was purified by amylose affinity chromatography, as described in the pMAL protein fusion and purification manual (NEB). All subsequent purification steps were performed on ice or at 4°C. The purified protein concentration was determined by the Bradford protein assay (Bio-Rad, Hercules, CA, USA). The quality and quantity of the recombinant proteins at different purification steps were evaluated by 12% sodium dodecyl sulfate-polyacrylamide electrophoresis (SDS-PAGE) under reducing conditions.
The antibacterial activity of AcLysC and AcLysG-like recombinant proteins were analyzed against four gram-negative (Edwardsiella tarda, Photobacterium damselae subsp . piscicida, Vibrio anguillarum, and V. harveyi) and two gram-positive (Micrococcus luteus and Streptococcus parauberis) using a previously reported turbidimetric assay (Xue et al., 2004) in a buffer system at pH 4.0 (sodium acetate–acetic acid, 50 mM) and 25°C. Initially, all the bacterial cultures were grown up to 0.4–0.5 at OD450 in relevant media. A portion of 150 μL from the bacterial suspensions was mixed with 50 μL of each recombinant protein (500 ng/μL) in a 96-well plate. The initial OD450 of the mixture was measured immediately using a microtiter plate reader (Multiskan EX, Thermo Scientific), and the final OD450 was measured after 30 min of incubation at 25°C. MBP (500 ng/μL) and standard hen egg white lysozyme (HEWL, 500 ng/μL; Biosesang, Seongnam, Korea) were used as negative and positive controls, respectively. The relative antibacterial activity of each recombinant protein in terms of lytic activity against the bacterial cell wall was calculated as the difference between the initial (ODi) and final (ODf) absorbance. The assay was performed in triplicates to increase the reliability of the results.
The lytic activity of AcLysC and AcLysG-like was investigated against M. luteus under a range of pH and temperature conditions using a turbidimetric assay (Xue et al., 2004). M. luteus has commonly been used as a standard bacterial strain to study antibacterial activity and has proven to be highly susceptible to lysozyme-mediated killing (Ganz et al., 2003). Bacterial suspensions (0.8 mg/mL) of M. luteus (Sigma-Aldrich) were prepared in different buffers with pH ranging from 3.0 to 10.0. Buffers were prepared with sodium acetate–acetic acid (50 mM, pH 3.0–5.0), Na2HPO4–NaH2PO4 (50 mM, pH 6.0–8.0), and boric acid–NaOH (50 mM, pH 9.0–10.0). Antibacterial assays using bacterial suspensions in each buffer were conducted as described in section 2.8 with respective controls. To determine the temperature-dependence of the lytic activity of recombinant proteins, the bacterial suspension was diluted with a buffer at pH 4, and the assay was carried out at a temperature ranging from 10°C to 45°C, with intervals of 5°C. The effects of pH and temperature on the lytic activity of each recombinant protein were calculated as the difference between ODi and ODf absorbance values. All assays were conducted in triplicate.
A bacterial suspension of M. luteus (0.8 mg/mL) was rinsed twice with phosphate buffer (pH 7.4) using two rounds of resuspension followed by centrifugation. Then, the bacterial pellet was mixed with either 500 μL of AcLysC (500 ng/μL), AcLysG-like (500 ng/μL), MBP (500 ng/μL), or molecular biology grade water. All mixtures were incubated at 25°C for 30 min and centrifuged at 16,200×g for 1 min. Each bacterial pellet was fixed in 0.1 M phosphate buffer containing 2% glutaraldehyde (pH 7.4) at room temperature for 1 h. Pellets were then dehydrated through a series of ethanol solutions (50%, 70%, 90%, 95%, and 100%) for 15 min each. Subsequently, samples were dried using liquid CO2, followed by platinum coating using a sputter coater (Quorum Technologies, Laughton, UK). The observations were made using a MIRa3 TESCAN SEM (TESCAN, Brno, Czech Republic) at 5 kV.
Results
As depicted in Supplementary Fig. S1A, the ORF of AcLysC consisted of 429 bps, encoding for a 142 amino acid sequence with a predicted N-terminal signal peptide (Met1-Ala17). Furthermore, the isoelectric point (pI) and calculated molecular weight of AcLysC were 8.2 pI and 14.03 kDa, respectively. AcLysC shared the highest identity (98.6%) and similarity (98.6%) with LysC from Amphiprion ocellaris, followed by LysC from P. olivaceus (Supplementary Table S1). AcLysG-like contained an ORF of 570 bp encoding a protein sequence of 189 amino acids (Supplementary Fig. S1B). The pI and calculated molecular weight of AcLysG-like protein were 6.51 pI and 20.48 kDa, respectively. The AcLysG-like shared the highest identity (84.6%) and similarity (88.2%) with its homolog from A. ocellaris (Supplementary Table S2).
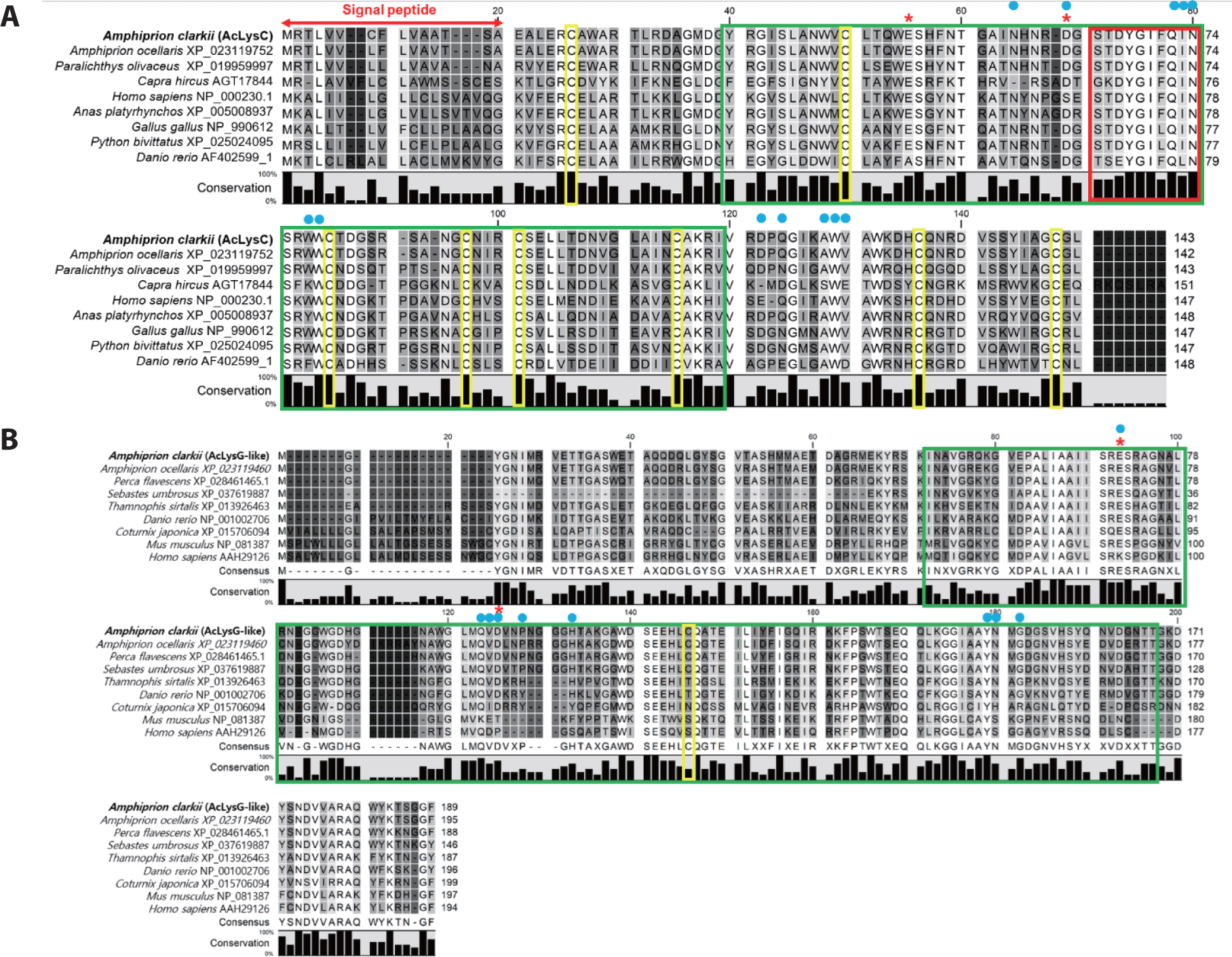
Prediction of domain organization demonstrated that both sequences contained a soluble lytic trans-glycosylase domain (SLT), catalytic residues, and sugar-binding sites (Fig. 1). According to the multiple sequence alignment, cysteine residues were mostly present in fish c-type lysozymes compared to fish g-like-type lysozymes. Furthermore, AcLysC shared a conserved catalytic motif with all its counterparts except for those from Capra hircus and Danio rerio (Fig. 1A). The AcLysG-like also contained conserved catalytic residues and sugar-binding sites, especially among fish orthologs (Fig. 1B).
To identify the molecular evolutionary relationship of AcLysC and AcLysG-like with its homologs, a phylogenetic tree was constructed using orthologs from fish, reptiles, birds, and mammals (Supplementary Fig. S2). According to the results, AcLysC was grouped in the clade encompassing fish c-type lysozyme and closely related to the c-type lysozyme from A. ocellaris. Similarly, AcLysG-like was clustered together with the fish g-like-type lysozymes and was closely related to its counterpart from A. ocellaris.
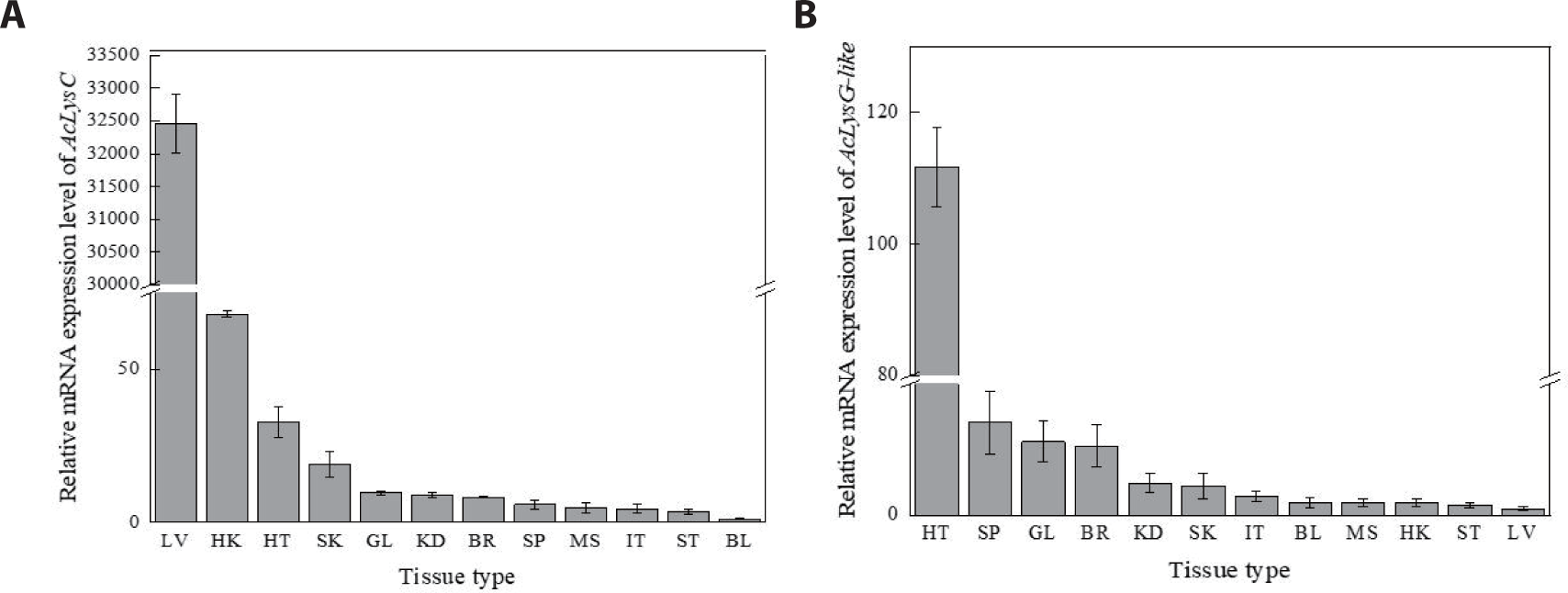
AcLysC and AcLysG-like were expressed in each selected tissue of healthy fish (Fig. 2). According to the tissue distribution of AcLysC (Fig. 2A), the highest expression was observed in the liver, followed by the head kidney, heart, and skin, while the lowest expression was observed in blood cells. The most prominent expression of AcLysG-like was observed in the heart, followed by the spleen, gill, and brain (Fig. 2B).
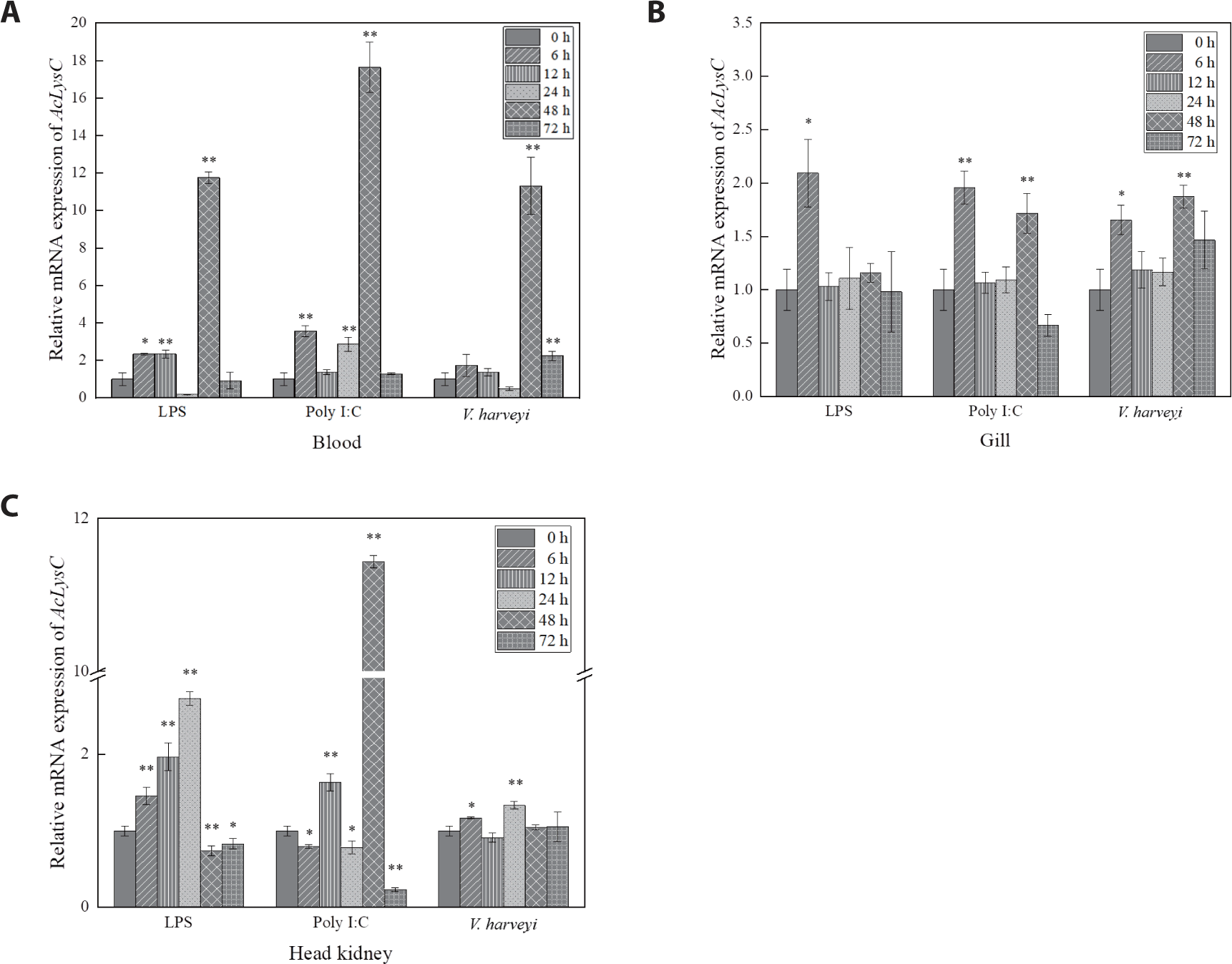
AcLysC and AcLysG-like expression after the immune stimulation with LPS, Poly I:C, and V. harveyi were investigated over 0–72 h p.i. in blood cells, gill, and head kidney (Figs. 3 and 4). The expression level of AcLysC in blood cells against each immune stimulant reached the peak (late-stage response) at 48 h p.i. In gills, the basal transcript level of AcLysC was remarkably upregulated against all stimuli at 6 h p.i. Additionally, significant upregulation of AcLysC was also observed against Poly I:C and V. harveyi at 48 h p.i. In head kidney tissues, AcLysC expression was significantly (p < 0.05) elevated relative to its basal level in response to each immune stimulation at multiple p.i. time points. In this regard, expressional inductions against LPS and Poly I:C treatments were observed at all p.i. time points (Fig. 3C).
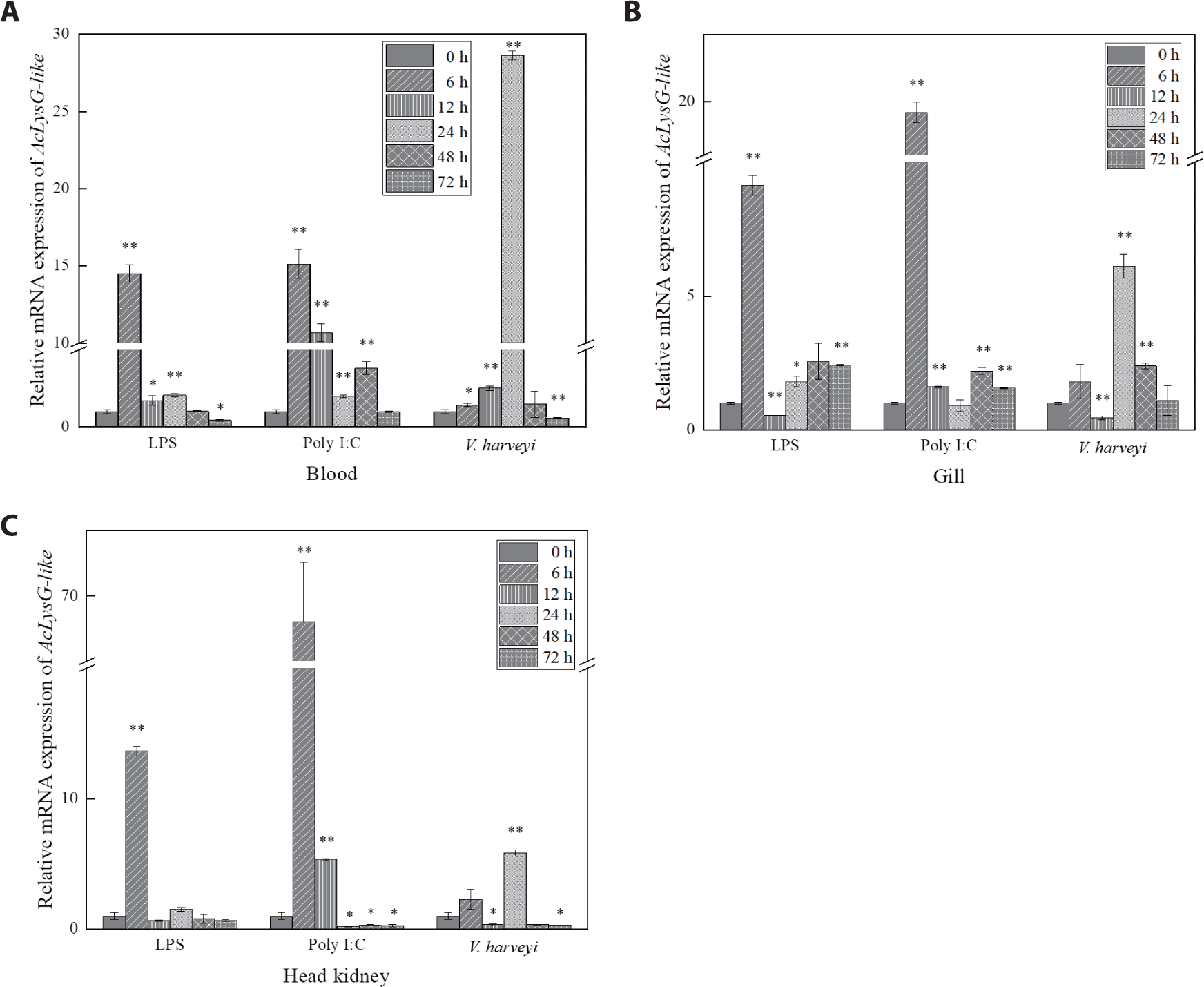
As depicted in Fig. 4, expressional inductions of AcLysG-like upon LPS and Poly I:C treatments were observed in gills and the head kidney at early phase p.i. (6 h p.i.), contrasting to what was observed for AcLysC in the same tissues. However, upon the experimental infection with V. harveyi, the expression of both AcLysC and AcLysG-like peaked at late phase p.i. (24–48 h p.i.).
AcLysC and AcLysG-like were overexpressed as soluble recombinant fusion proteins with MBP by IPTG induction at 25°C for 8 h in E. coli ER2523. We then purified fusion proteins by affinity chromatography. The eluted recombinant fusion AcLys proteins (rAcLys) were separated and analyzed using 12% SDS-PAGE. The expected molecular weights of rAcLysC and rAcLysG-like were approximately 56.5 kDa and 62.9 kDa, respectively, as the molecular weight of MBP is approximately 42.5 kDa. Intact single bands corresponding to each expected size were observed with SDS-PAGE on their respective lanes (Supplementary Fig. S3), indicating the sufficient purity of proteins.
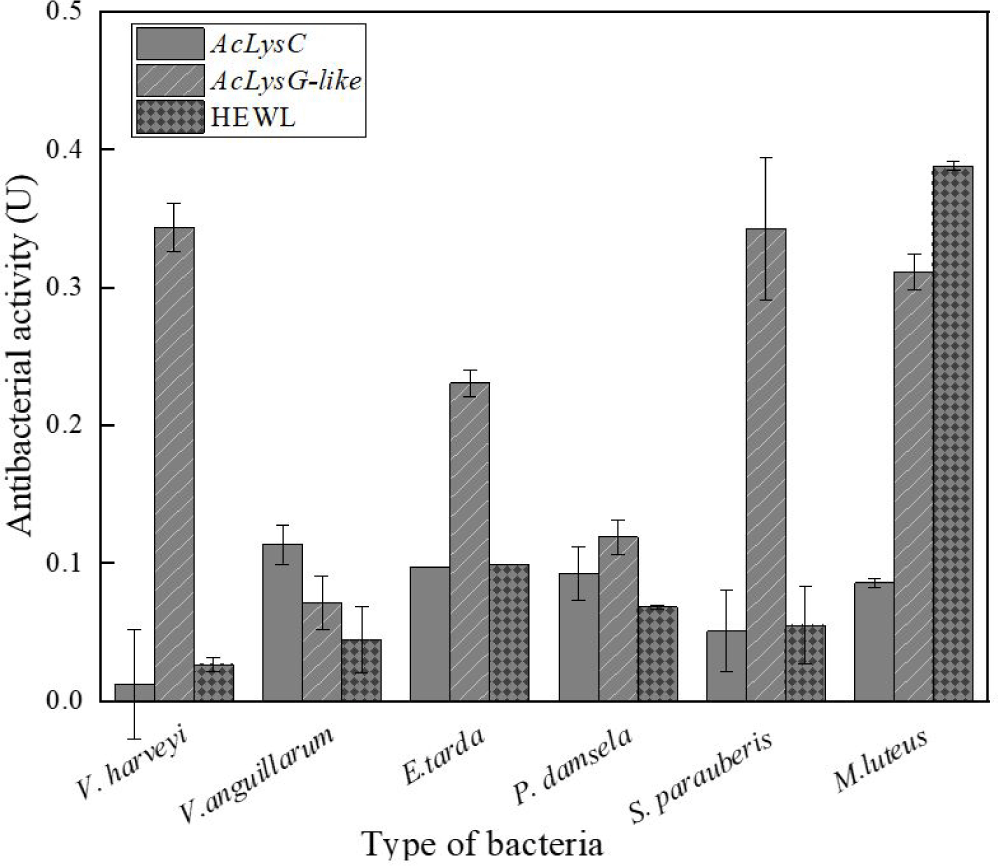
As shown in Fig. 5, the antibacterial activities of AcLysC and AcLysG-like were observed against different marine bacterial pathogens. Particularly, AcLysG-like had more pronounced antibacterial activity against most bacteria compared to that of AcLysC and HEWL. AcLysC exhibited lower antibacterial activity against most bacteria used in the study compared to the positive control HEWL. However, AcLysC showed its highest antibacterial activity against V. anguillarum, and HEWL showed more prominent antibacterial activity against M. luteus.
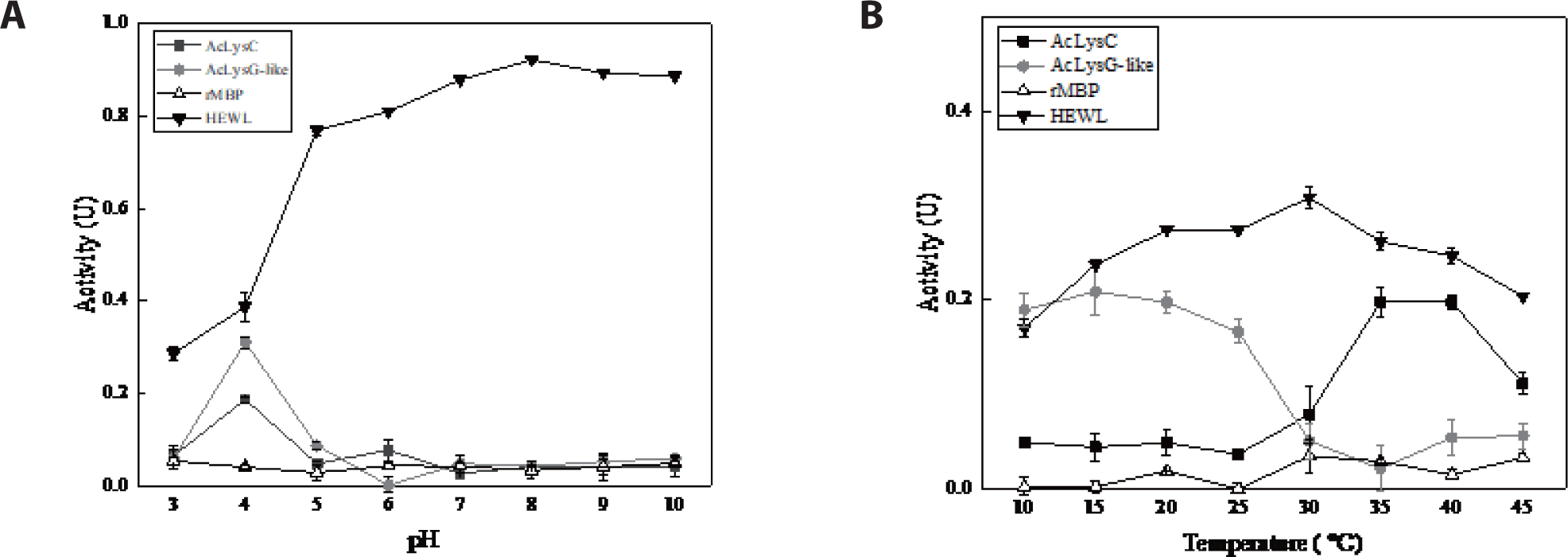
To investigate the modulation of antibacterial activity of AcLysC and AcLysG-like with pH and temperature, the bactericidal activity of recombinant fusion proteins was assessed under a range of pH and temperatures against M. luteus, with MBP and HEWL as controls. Results showed that both AcLysC and AcLysG-like exhibit their highest bacteriolytic enzymatic activity at pH 4.0. The highest enzymatic activity of HEWL (positive control) was observed at pH 8 (Fig. 6A). Additionally, AcLysC was found to be noticeably active at moderately high temperatures (35°C–40°C), while AcLysG-like was active at lower temperatures (10°C–15°C) (Fig. 6B). HEWL showed the highest activity at 30°C, whereas no significant activity was detected for MBP under tested pH and temperature conditions.
To observe the morphological changes that occurred due to the bacteria lytic activities of the two proteins, the cell surface of M. luteus was filmed using a scanning electron microscope (SEM; MIRa3 TESCAN). Generally, naïve M. luteus cells have a smooth surface with a spherical (coccus) shape. Expectedly, both the H2O- and MBP-treated samples showed clear and smooth cell surfaces (Fig. 7A and 7B). In contrast, M. luteus treated with HEWL (Fig. 7C), AcLysC (Fig. 7D), and AcLysG-like (Fig. 7E) showed wrinkled, shrunken, and fractured cell walls, evidencing cell lysis.

Discussion
Lysozyme is one of the most important antibacterial proteins involved in host innate immune mechanisms of the host. It breaks the peptidoglycan layer of the cell wall, thus promoting the lysis of bacterial cells (Jollès & Jollès, 1984). Furthermore, lysozyme can be found in many organisms, including bacteriophages, microbes, plants, invertebrates, and vertebrates. It shows antibacterial activity against various bacteria and controls the release of pathogen-associated molecular patterns (PAMPs), hormones, and (Whang et al., 2011; Zhao et al., 2011) involved in the regulation of immune responses (Ragland & Criss, 2017).
According to the in silico analysis, both AcLyC and AcLyG-like contain the SLT domain, which plays a critical role in the hydrolysis of β-1,4-glycosidic bonds between N-acetylmuramic acid and N-acetylglucosamine of the peptidoglycan layer in the cell wall (Whang et al., 2011). In addition, AcLysC harbors a conserved sugar-binding site, cysteine residues, and catalytic residues in common with those found in other species. The AcLysC protein sequence comprises a high content of well-conserved cysteine residues, which are known to form four intra-disulfide bonds that might provide high stability and osmolarity against seawater and proteases (Ito et al., 1999). These disulfide bridges are considered to remarkably affect the catalytic function, secretion processes, and structural stabilization by maintaining the molecular integrity and elliptical shape of the protein (Kato et al., 1981). Additionally, a signal peptide sequence in AcLysC can help secrete the protein extracellularly, as observed for most known c-type lysozymes (Cai et al., 2019). Extracellular lysozymes also break down the peptidoglycan layer into soluble fragments, enabling PAMP recognition by nucleotide-binding oligomerization domain-containing protein receptors in mucous epithelial cells, which stimulate the chemical maintenance and secretion of immune activation factors in macrophages (Ragland & Criss, 2017). On the other hand, the AcLysG-like protein sequence lacks disulfide bonds (only one cysteine residue) and a signal peptide. Therefore, AcLysG-like can be considered an intracellular protein. Moreover, previous reports on fish such as Atlantic cod (Gadus morhua L.) (Larsen et al., 2009), olive flounder (P. olivaceus) (Hikima et al., 2001), and orange-spotted grouper (E. coioides) (Wei et al., 2012) confirmed the absence of cysteines in g-type lysozymes. However, the number of cysteine residues in g-type lysozymes varies depending on the species (Kawamura et al., 2006; Kawamura et al., 2008).
According to a previous study, the lysozyme content of fish organs can differ based on the species (Subbotkina & Subbotkin, 2003). Furthermore, lysozyme showed higher expression in the kidney, spleen, and liver compared to other tissues of many fish species (Gao et al., 2012; Jiménez-Cantizano et al., 2008; Wang et al., 2016). According to the tissue-specific expression patterns observed in this study, the highest AcLysC mRNA expression level was detected in the liver, the main organ involved in metabolism, detoxification, and hematopoiesis in fish (Rui, 2014). Lysozymes play an additional role in nutrient absorption in animals (Callewaert & Michiels, 2010). For instance, rabbits have unique behavior, “coprophagia,” a nutritional strategy to absorb large amounts of lysozyme from soft feces (Cámara & Prieur, 1984). Fish can be continuously exposed to various pathogens during feeding. Thus, the liver and its secretions, such as the bile, could contain a high amount of lysozyme as a defensive mechanism. Furthermore, fish c-type lysozymes showed high mRNA levels depending on various factors, such as stress, intensity, and diseases (Caruso et al., 2002). Therefore, it is likely to observe pronounced expression of AcLysC in liver tissues.
In contrast, g-type lysozymes are specialized to play a distinct role in antibacterial activity (Saurabh & Sahoo, 2008). The highest expression level of AcLysG-like was observed in the heart, followed by that in the spleen and gills. A similar mRNA expression pattern was earlier detected in olive flounder (P. olivaceus) (Hikima et al., 2001). The heart and spleen are reportedly involved in the clearance of blood-borne substances, particularly in some fish species like G. morhua, and endothelial cells of the heart were found to be involved in the endocytosis of circulating substances. In contrast, gills can act as a barrier to the entry of pathogens and are enriched with leucocytes which can contribute to the local immune responses (Press & Evensen, 1999). Thus, intracellular lysozymes such as AcLysG-like seemingly exert bacteriolytic functions to protect the host from infections.
Based on the temporal mRNA expression, the defensive roles of AcLysC and AcLysG-like against LPS, Poly I:C, and V. harveyi were investigated in blood cells, gill, and head kidney tissues. Unlike those of mammals, fish red blood cells are nucleated and can trigger immune responses against immune stimulants (Morera & MacKenzie, 2011). However, peripheral white blood cells, such as macrophages and neutrophils, which act as first-line barriers to pathogenic infections, abundantly express lysozymes. Macrophages and neutrophils engulf pathogens and kill them via lysozyme-mediated degradation (Callewaert & Michiels, 2010). Therefore, lysozymes in fish blood cells may play a vital role in combating infections. Fish gills are more susceptible to infectious pathogens due to their continuous exposure to ambient water that circulates during respiration. The fish head kidney is analogous to the adrenal gland of mammals and is a leukocyte-rich immune organ that subsequently activates several other immune mechanisms in other tissues (Geven & Klaren, 2017; Paulsen et al., 2003). Expectedly, the expression of AcLysC and AcLysG-like was significantly modulated by immune stimulants in these tissues.
An endotoxic immune modulator, LPS, is known to induce lysozyme production through Toll-like family receptors or cytokines (Jiménez-Cantizano et al., 2008). LPS stimulation induced lysozyme upregulation in brill (Scophthalmus rhombus) (Yildiz, 2006) and yellow catfish (Pelteobagrus fulvidraco) (Liu et al., 2016). Similarly, significant upregulation of AcLysC and AcLysG-like mRNA levels was observed in the blood, gill, and head kidney tissues after LPS treatment.
Poly I:C is a synthetic compound that, like a double-stranded RNA virus, stimulates an antiviral immune response. Previous studies have provided evidence for the antiviral activity of lysozyme (Lee-Huang et al., 1999; Mai & Wang, 2010; Takahashi et al., 2018; Zhang et al., 2008). Lysozymes in chicken egg white, human milk, and human neutrophils have been shown to degrade the viral RNA of human immunodeficiency virus (Lee-Huang et al., 1999). Given this background, the observed inductive expressional responses of AcLysC and AcLysG-like upon Poly I:C treatment suggest their potential role in antibacterial and antiviral defense in the piscine innate immune system.
V. harveyi is a gram-negative marine bacterium that is associated with a broad spectrum of diseases in fish (Zhang et al., 2020). Although gram-negative bacteria are less vulnerable to lysozymes, basal expression of both AcLysC and AcLysG-like were upregulated after V. harveyi challenge. However, AcLysG-like induced its basal expression with a higher fold than that of AcLysC post-experimental infection with V. harveyi. A similar pattern was observed in grass carp when challenged with Aeromonas hydrophila (Ye et al., 2010) . Therefore, AcLysG-like might be more sensitive against V. harveyi than AcLysC in host immune defense. In contrast to the AcLysG-like responses against LPS and Poly I:C, upregulation was detected at a later phase against V. harveyi infection. This observation can be attributed to the fact that the piscine innate immune system requires more time to respond to the live pathogens compared to PAMPs such as LPS and Poly I:C. Live pathogens may have additional patterns deriving from the strategies that are used to invade, manipulate, replicate within, or spread among their hosts (Vance et al., 2009).
Several studies on animals such as barramundi (Lates calcarifer) (Fu et al., 2013), silkworm (Bombyx mori) (Chen et al., 2018), Farrer’s scallop (Chlamys farreri) (Zhao et al., 2007), grass carp (Ctenopharyngodon idellus) (Ye et al., 2010), and Chinese perch (Siniperca chuatsi) (Sun et al., 2006) have shown that recombinant lysozymes exert promising antimicrobial activity against bacterial pathogens. The selected bacterial species in this study such as E. tarda, P. damselae subsp. piscicida, V. anguillarum, V. harveyi, and S. parauberis are well-known disease-causing pathogens of mariculture fish (Austin & Zhang, 2006; Blazer, 1988; Currás et al., 2002; Rivas et al., 2013). According to our results, AcLysG-like demonstrated more pronounced antibacterial activity against most bacterial pathogens used in the study. Further, it showed profound activity against both gram-negative and gram-positive bacteria compared to AcLysC. Lysozymes are known to show stronger bacteriolytic activity against gram-positive than gram-negative bacteria (Saurabh & Sahoo, 2008; Silvetti et al., 2017) but they can also extend activity against gram-negative bacteria through denaturation, chemical modifications, or by combining themselves with other preservatives (Silvetti et al., 2017). Apart from that, it is reported that g-type lysozymes are able to bind to both gram-positive and negative bacterial cell wall components (LPS (Lipopolysaccharide), PGN (Peptidoglycan), and LTA (Lipoteichoic acid)) without significant differences (Liu et al., 2022). Moreover, it is speculated that g-like type lysozymes are able to cause more denaturation or chemical modifications in bacteria than c-type lysozyme, which contains numerous cysteine residues with stable disulfide bonds (Clarke et al., 2000; Kawamura et al., 2008). AcLysC showed relatively low antibacterial activity against V. harveyi. One possible explanation behind this observation is the antibacterial resistance of this bacterium to lysozyme either by altering the cell wall structure or through covalent binding with other polymers of the peptidoglycan cell wall (Bera et al., 2006; Callewaert et al., 2012; Raymond et al., 2005). Additionally, we observed that AcLysG-like showed different antimicrobial activity against V. anguillarum and V. harveyi. Several reports suggest that g-type lysozymes show different rates of antimicrobial activity with bacterial species from the same genus such as V. tapetis and V. anguillarum (Nilojan et al., 2017), and S. aureus and S. iniae (Wei et al., 2014). The reason might be that g-type lysozyme has a different binding ability to bacteria or bacteria cell wall components. Due to these different binding abilities, g-type lysozymes selectively kill gram-negative bacteria in a non-enzymatic mode by damaging bacterial membranes (Luo et al., 2021). Collectively both lysozymes can be proposed as potential players in the host antibacterial defense in A. clarkii. The differential activities of two AcLys against the same pathogen may reflect the pathogenetic differences underlying bacterial diseases or the consequences of distinct infection routes (Wang et al., 2013). Previous studies have suggested that g-type lysozyme may be induced for defense against invading pathogens, whereas c-type lysozyme might be the major molecule for housekeeping defense under normal conditions (Ye et al., 2010). This notion can also be inferred by comparing the antibacterial activities observed for rAcLysG-like with rAcLysC in this study.
Recombinant AcLysC was noticeably active at lower pH (pH 4) and moderately high temperatures (35°C–40°C). Similar observations have been reported for disk abalone (Haliotis discus discus) (Umasuthan et al., 2013) and Mediterranean mussel (Mytilus galloprovincialis) (Wang et al., 2013), showing that their c-type lysozymes have pronounced activity at low pH. Like AcLysC, AcLysG-like also showed its optimum activity at low pH (pH 4). Previously, g-type lysozymes were reported to be active at low pH in black rockfish (Sebastes schlegelii) (Nilojan et al., 2017), big-belly seahorse (H. abdominalis) (Ko et al., 2016), rock bream (Oplegnathus fasciatus) (Whang et al., 2011), and disk abalone (H. discus discus) (Bathige et al., 2013).
The optimum temperature of enzymes was shown to depend on the number of disulfide bonds (Li et al., 1998; Yin et al., 2015). The AcLysC sequence contains eight cysteine residues. However, AcLysG-like contains only one cysteine residue. According to our observation, rAcLysC, which can potentially form more disulfide bonds, likely demonstrate its optimal activity at a higher temperature range compared to rAcLysG-like.
Scanning electron microscope examination of M. luteus following rAcLys treatments evidenced morphological changes in the bacterial cell wall. The surface changes of M. luteus observed after the treatments reinforced the detected antibacterial activities of rAcLys using the turbidimetric assay.
Conclusion
In summary, AcLysC and AcLysG-like lysozyme sequences of A. clarkii were conserved relative to those of other homologs and were closely related to A. ocellaris orthologs. The highest tissue-specific mRNA levels of AcLysC and AcLysG-like were observed in the liver and the heart, respectively. Furthermore, AcLysC and AcLysG-like were upregulated in the gill, head kidney, and blood cells following immune stimulation. Recombinant AcLysC and AcLysG-like proteins showed antibacterial activity against all tested fish pathogens with the highest lytic activity at pH 4. The SEM observations provided conclusive evidence of the lytic activity of AcLysC and AcLysG-like on the cell wall of M. luteus. Altogether, the findings of this study suggest that AcLysC and AcLysG-like play a crucial role in the innate immune system of A. clarkii by defending against invading microbial pathogens.
