Background
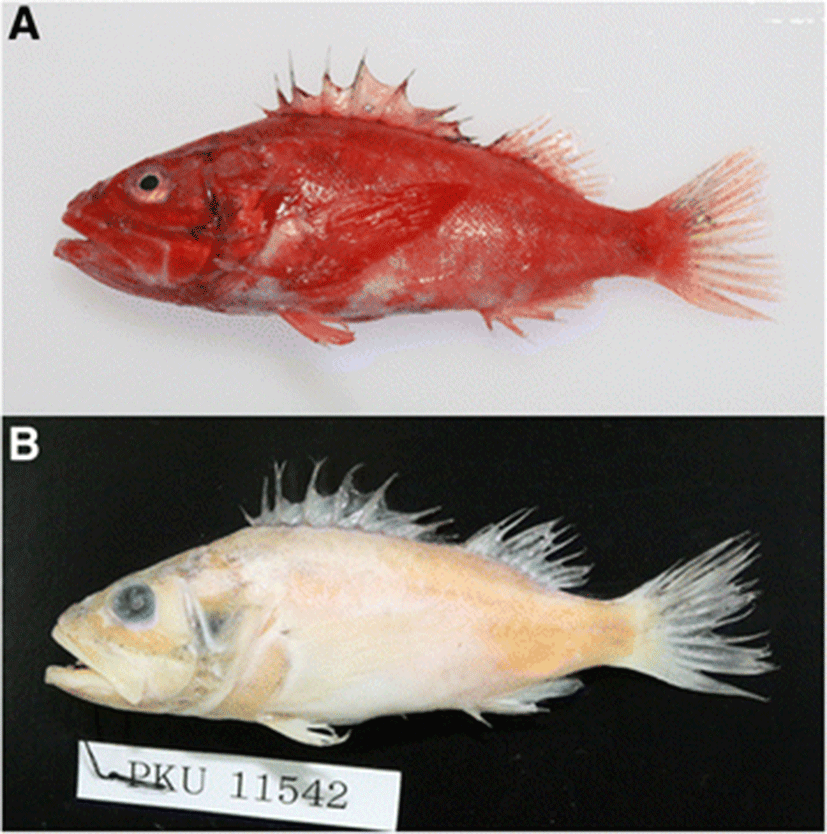
The family Scorpaenidae, in the suborder Scorpaenoidei, contains 418 species and 56 genera throughout the world, with 45 species and 17 genera in Korea (Han et al. 2011; Nelson 2006). This family is widely distributed along all coasts of the world, with most species in the Indian and Pacific Oceans (Nelson 2006). The species of Scorpaenidae are characterized by a strong spine and a well-developed ridge on the head, 3–5 preopercular spines, and a dorsal fin that is usually single with a V-shaped notch (Kim et al. 2005). The genus Setarches contains three species throughout the world: Setarches guentheri Johnson, 1862, Setarches longimanus (Alcock, 1894), and Setarches armata (Fowler, 1938), and the former two of which occur in Japan (Froese & Pauly 2015; Nakabo & Kai 2013). However, no occurrence of the genus Setarches has been reported in Korea. In this study, we confirmed that the specimens collected from Busan and the coastal waters of Jeju Island, Korea, were S. longimanus using morphological and molecular methods, and we provide a morphological description of these specimens as the new record based on the specimens (Fig. 1).
Methods
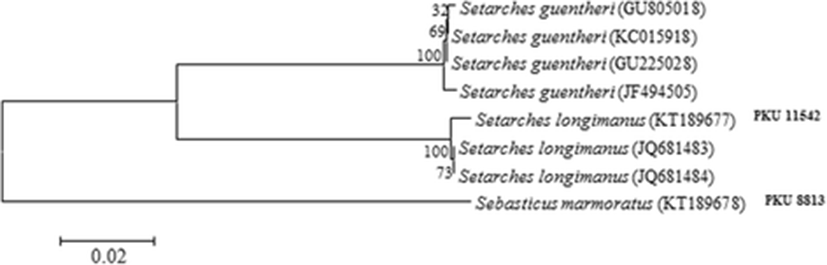
The two specimens belonging to S. longimanus were collected for the first time from Busan and the coastal waters of Jeju Island, Korea, on January and November, 2014, and were fixed in 10 % formalin and preserved in 70 % ethanol. Counts and measurements were made according to the methods of Eschmeyer (Eschmeyer 1965) and Hubbs and Lagler (Hubbs & Lagler 2004) (body parts were measured to the nearest 0.1 mm with Vernier calipers). The number of vertebrae and all fin rays were counted from radiographs (Sehwa Medical System SMS-CM, Korea). The specimens of S. longimanus are deposited at the Ichthyological Laboratory, Pukyong National University (PKU) and Jeollabuk-do Fisheries Research Institute (JBFRI), Korea. Genomic DNA was extracted from muscle tissues using a DNA Extraction Kit (Bioneer Trade Co. LTD, Korea). The specimens were performed using VF2 (5’-TCA ACC AAC CAC AAA GAC ATT GGC AC-3’) and FishR2 (5’-ACT TCA GGG TGA CCG AAG AAT CAG AA-3’) primers, which amplify the mitochondrial DNA cytochrome oxidase subunit I (COI) (Ivanova et al. 2007; Ward et al. 2005). The polymerase chain reaction (PCR), purification, and sequencing used to modify the method of (Ward et al. 2005). The sequences were aligned using ClustalW (Thompson et al. 1994) in BioEdit ver. 7 (Hall 1999). The sequences of S. guentheri (GU225028; GU805018; JF494505; KC015918) and S. longimanus (JQ681483; JQ681484) from the National Center for Biological Information database, and Sebastiscus marmoratus (PKU 8813) was used as an outgroup. Genetic distances were calculated using the Kimura two-parameter method (Kimura 1980) in MEGA 5 (Tamura et al. 2011). A neighbor-joining (NJ) tree was constructed with the Kimura two-parameter method (Kimura 1980) and 10,000 bootstrap replications using MEGA 5 (Tamura et al. 2011).
Results and Discussion
(New Korean name: Ma-su-gam-peng-sok)
Setarches Johnson, 1862: 177 (type species, Setarches guentheri Johnson, 1862)
Bathysebastes Döderlein in Steindachner and Döderlein, 1884: 207 (Bathysebastes albescens Döderlein, 1884)
Lythrichthys Jordan and Starks, 1904: 140 (Lythrichthys eulabes Jordan and Starks, 1904)
Scorpaenopsella Fowler, 1938: 67 (Scorpaenopsella armata Fowler, 1938)
Scorpaenella Fowler, 1938: 67 (Scorpaenella cypho Fowler, 1938)
Body moderately compressed; head scaleless, except for cheek and behind eye; maxilla without keel; preopercular with five spines; anterior lower infraorbital spine subequal to the two posterior spines; when fresh, body and head are red, and the whole digestive duct is black or dark gray; swim bladder well developed (Nakabo & Kai 2013; Eschmeyer & Collette 1966).
(New Korean name: Bul-geun-ma-su-gam-peng) (Table 1, Fig. 1)
Morphological characters | Present study | Alcock (1894) | Eschmeyer and Collette (1966) | Yamada et al. (2009) | Nakabo and Kai (2013) |
|---|---|---|---|---|---|
No. specimens | 2 | 1 | 24 | - | - |
Standard length (mm) | 115–176.3 | 156.7 | 30–162 | - | - |
Counts | |||||
Dorsal fin rays | XI,10 | XII, 11 | XI–XIII, 9–11 | XII–XIII, 9–10 | XI–XIII, 9–11 |
Anal fin rays | III, 5 | III, 5 | III, 4–6 | III, 5 | II–III, 4–6 |
Pectoral fin rays | 19–21 | 22 | 21–23 | 21–22 | 20–23 |
Pelvic fin rays | I, 5 | I, 5 | - | - | I, 5 |
Vertebrae | 24 | - | - | 25 | - |
Lateral line scales | 26–28 | - | - | - | - |
Gill rakers | 3 + 9–10 | - | - | 2 + 9–10 | - |
In % standard length | |||||
Head length | 38.5–42.0 | - | - | - | - |
Snout length | 13.0–13.3 | - | - | - | - |
Orbit diameter | 7.2–8.3 | - | - | - | - |
Interorbital width | 11.3–11.6 | - | 9–12 | - | - |
Body depth | 28.4–31.8 | - | 31–38 | - | - |
Caudal peduncle depth | 9.9–10.0 | - | - | - | - |
Upper jaw length | 19.7–21.6 | - | - | - | - |
Predorsal fin length | 36.3–38.4 | - | - | - | - |
Preanal fin length | 69.8–70.1 | - | - | - | - |
Pectoral fin length | 30.5–31.0 | - | - | - | - |
Pelvic fin length | 18.3–20.1 | - | - | - | - |
Length of dorsal base | 52.5–53.7 | - | - | - | - |
4th dorsal fin spine length | 10.3–11.1 | - | - | - | - |
1st anal fin spine length | 4.8–6.1 | - | - | - | - |
Caudal length | 21.6–22.8 | - | - | - | - |
Lioscorpius longiceps Günther, 1880: 52 (type locality: Romblon Island, Philippines).
Lioscorpius longiceps longimanus Alcock, 1894: no page number, Pl. 10, Fig. 3 (Andaman Sea).
Setarches longimanus: Matsubara, 1943: 372–385 (Japan) Eschmeyer and Collette, 1966: 356–357; Nakabo and Kai in Nakabo, 2013: 685.
PKU 11542, one specimen, 115 mm in SL, off Jeju Island, Korea, on January 6, 2014, collected by W.J. Lee; JBFRI 1198, one specimen, 176.3 mm in SL, Busan, Korea, on November 21, 2014, collected by D. S. Joo.
Dorsal fin rays XI, 10; anal fin rays III, 5; pectoral fin rays 19–21; pelvic fin rays I, 5; lateral line scales 26–28; gill rakers 3 + 9–10; vertebrae 24 (Table 1).
The measurements are presented as a percentages of standard length (SL): head length, 38.5–42; snout length, 13–13.3; orbit diameter, 7.2–8.3; interorbital width, 11.3–11.6; body depth, 28.4–31.8; caudal peduncle depth, 9.9–10; upper jaw length, 19.7–21.6; predorsal fin length, 36.3–38.4; preanal fin length, 69.8–70.1; pectoral fin length, 30.5–31; pelvic fin length, 18.3–20.1; length of dorsal fin base, 52.5–53.7; fourth dorsal fin spine length, 10.3–11.1; first anal fin spine length, 4.8–6.1; caudal fin length, 21.6–22.8.
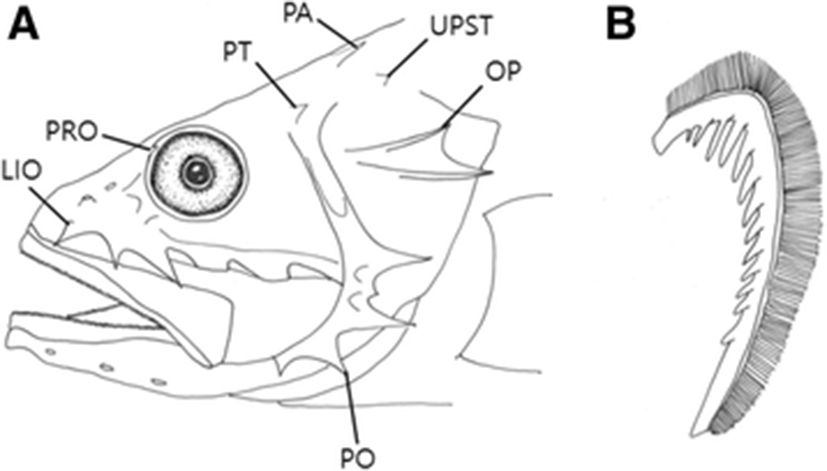
Body oval and moderately compressed (Fig. 1). Head large (38.5–42 % of SL). Snout short and blunt (13–13.3 % SL). Lower jaw protrudes anteriorly; small conical teeth on both jaws in many rows; posterior margin of the maxilla at right angle, reaching to the posterior margin of the orbit; maxilla without keel; ventral side of the lower jaw with three pairs of sensory pores. Two pairs of nostrils, located in front of the orbit; anterior nostril with short tube; posterior nostril circular. Eyes circular and close to the dorsal margin of the head. Spines developed on the head: preocular with one spine; pterotic with one small spine; parietal spine long, well developed, and sharp; infraorbital with three spines, anterior lower infraorbital spine reaching to the maxilla; preopercular with five spines, second spine shorter than the first and third; opercular with two spines, upper spine shorter than the lower spine; upper posttemporal spine above the opercular spine (Fig. 2a). The posterior tip of the opercular region extends to the fourth spine of the dorsal fin. Dorsal fin single; dorsal fin base long; origin of dorsal fin vertically above the origin of the pelvic fin; V-shaped notch in front of the last dorsal spine. Pectoral fin long; pectoral fin begins under the third spine of the dorsal fin and extends to the origin of the anal fin. Pelvic fins located below the pectoral fins; pelvic fin 1/3 the length of the pectoral fin. Origin of the anal fin below the first dorsal fin ray, located in front of the anus. Caudal fin truncated. Body covered with cycloid scales; head scaleless, except for the cheek and behind the eye. Lateral line single, begins at the upper tip of the gill opening and extends to the base of the caudal fin.
When fresh, the body and head are red. The operculum, nape, and pectoral fin base have small dark spots. After fixation in formalin, both the body and head are light yellow, and small dark spots are distributed on them.
Busan and Jeju Island in Korea (present study), Japan (Nakabo & Kai 2013), China (Chinese Academy of Fishery Science (CAFS 2007), western Pacific Ocean (Eschmeyer & Collette 1966; Gloerfelt et al. 1984; Kulbicki et al. 1994), southward to Taiwan (Shao et al. 2008), Andaman Sea (Rajan et al. 2011), and Arabian Sea (Nakabo & Kai 2013).
The present specimens were collected from Busan and Jeju Island, Korea, and identified as belonging to the genus Setarches, based on the following features: body moderately compressed; body and head are red; head scaleless, except for cheek and behind the eye; maxilla without keel; anterior lower infraorbital spine subequal to the two posterior spines. These specimens were assigned to S. longimanus based on following features: second preopercular spine shorter than the first and third; interorbital width 9–12 % SL (11.3–11.6 % SL in this study) (Nakabo & Kai 2013; Eschmeyer & Collette 1966). When compared with the original description (Alcock 1894), most counts were well corresponded to our specimens (Table 1), but our specimens differed slightly from that of Yamada et al. (2009) in the number of gill rakers, pectoral fin rays and vertebrae (Table 1; Fig. 2b). To confirm the taxonomic status of the specimen, we analyzed 587 base pairs of the mitochondrial DNA cytochrome c oxidase subunit I sequences. As a result, the sequences of our specimen (KT189677) almost corresponded to those of Chinese S. longimanus (genetic distance, d = 0.005), but considerably differed from those of S. guentheri (d = 0.120–0.124). Their relationship was also strongly supported by the neighbor joining tree with high bootstrap values (Fig. 3). Therefore, these subtle differences in some counts seem to be geographic variations within species. Most of the meristic characters in our specimens corresponded to those of S. guentheri and S. armata, but our specimens differed in their preopercular spines (second spine shorter than the first and third in S. longimanus vs. second spine subequal to or longer than the first and third in S. guentheri vs. second spine longer than third in S. armata) (Nakabo & Kai 2013; Eschmeyer & Collette 1966; Fowler 1938). Our specimens can be distinguished from S. guentheri in its interorbital width (9–12 % SL in S. longimanus vs. 7–9 % SL in S. guentheri) (Eschmeyer & Collette 1966), and from S. armata in its number of gill rakers (2 + 9–10 in S. longimanus vs. 6 + 12 in S. armata) (Yamada et al. 2009; Fowler 1938). We propose the new Korean genus name “Ma-su-gam-peng-sok”for the genus Setarches, and the Korean name of S. longimanus was already proposed by Yamada et al. (2009) as “Bul-geun-ma-su-gam-peng”, we followed the name for the species.
Conclusions
The two specimens belonging to S. longimanus were collected for the first time from Busan and the coastal waters of Jeju Island, Korea, on January and November, 2014. These specimens were assigned to S. longimanus based on following features: second preopercular spine shorter than the first and third; interorbital width 9–12 % SL (11.3–11.6 % SL in this study) An analysis of 562 base pair sequences of mitochondrial DNA cytochrome c oxidase subunit I showed that the sequences of our specimen (KT189677) almost corresponded to those of Chinese S. longimanus (genetic distance, d = 0.005), but considerably differed from those of S. guentheri (d = 0.120–0.124).








