Background
Anisakid nematodes (Nematoda: Anisakidae) infect animals belonging to almost all phyla and commonly found in aquatic vertebrates. Their life cycles involve various hosts at different levels in marine food webs. Crustaceans work as the first intermediate hosts; fish and cephalopods work as intermediate or paratenic hosts and marine mammals and fish-eating birds as definitive hosts (Mattiucci and Nascetti 2008). Since Van Thiel et al. (1960) reported that the third stage larvae (L3) of Anisakis from the Atlantic herring (Clupea harengus) can infect human, the number of clinical cases has been increasing worldwide, with the increase of knowledge about this parasite and improvement of diagnostics, as well as more globalized customs of eating raw marine fish (Cipriani et al. 2015).
The consumption of raw or undercooked fish harboring anisakid nematode L3 can lead to human infection (Audicana and Kennedy 2008). According to Fagerholm (1991), 10 genera exist in the family Anisakidae. However, the human infection is known to be caused frequently by the L3 of the genera Anisakis Dujardin, 1845 and less frequently by Pseudoterranova Krabbe, 1878 (Sakanari and McKerrow 1989). Of the genus Anisakis, A. simplex sensu stricto (s.s.) and A. pegreffii are recognized as zoonotic species causing anisakiasis in Asia and Europe, respectively (Mattiucci et al. 2013; Umehara et al. 2007). Thus, not all the nematode larvae found in fish are infective to human, and a clear diagnosis is important for proper epidemiological survey and further prevention of human infection.
Identification of anisakid nematodes has been traditionally conducted by morphological observation, but it is not always sufficient to make a clear identification at the species level because morphological characters of taxonomic significance in this group are very few, particularly for the larval stages lacking reliable diagnostic features at the species level (Mattiucci and Nascetti 2008). Moreover, morphological identification of nematodes is not always efficient when a huge number of the specimens have to be identified or the specimens are physically damaged and the morphological keys are missed. DNA-based diagnostic techniques such as polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) of internal transcribed spacer (ITS) region, direct sequencing of ribosomal DNA (rDNA), and mitochondrial DNA cytochrome c oxidase subunit 2 (mtDNA cox2) gene analysis have solved these constraints and have been widely used (Mattiucci and Nascetti 2008 and the references therein). Current taxonomy of the genus Anisakis based on these techniques revealed that there are two clades including nine species in this genus, and A. simplex, the most frequently encountered Anisakis species in human infection belongs to clade I (Mattiucci and Nascetti 2008).
The largehead hairtail or cutlassfish (Trichiuridae: Trichiurus species) distributes in circumtropical and temperate waters of the world and has been considered as one of the economically important fish species in the western North Pacific including Korea (Nakamura and Parin 1993). The annual catch of the largehead hairtail in Korea has been fluctuating between 50,000 and 90,000 t, after a sharp decrease in mid-1990 (Kim et al. 2011). Thus, importation of frozen fish from several countries has been increasing to meet the domestic demand (Cha and Kim 2009). Juveniles feed mostly on euphausiids, small pelagic planktonic crustaceans and small fishes, and adults feed mainly on fishes and occasionally on squids and crustaceans (Nakamura and Parin 1993). All of these food items are known to be the intermediate hosts of anisakid nematodes (Mattiucci and Nascetti 2008).
There have been many papers regarding anisakid nematodes isolated from Trichiurus species (Borges et al. 2012; Kong et al. 2015; Lee et al. 2009; Shih 2004; Umehara et al. 2010; Zhang et al. 2013). But, the identification of host species was not clearly conducted, and several scientific names (e.g., Trichiurus lepturus, Trichiurus haumela, Trichiurus sp.) have been indiscriminately used for the largehead hairtail in their studies. However, clear identification of host species, together with the information on their geographical distribution and feeding behavior are important to understand the occurrence of anisakid nematodes and predict the likelihood of infection in a given area. In particular, the largehead hairtails are often sold as fresh or frozen fillets, which are difficult to identify since most of the morphological features are no longer available. In this case, other techniques are necessary to identify these morphologically similar and closely related species in commercial products.
Although the largehead hairtail is commercially and economically important fish species in Korea, its anisakid nematode fauna has not been clearly investigated and only fragmentary information exists. Lee et al. (2009) reported A. pegreffii and Anisakis typica from several marine fish species including T. lepturus, but they did neither mention the origin of the fish nor the prevalence of infection in each host fish species. Moreover, there was no information in the microhabitat of anisakid nematodes in host fish. In this study, we investigated anisakid nematode fauna from the largehead hairtail caught from Korean waters. We collected nematodes in the largehead hairtail freshly caught from coastal waters around Jeju, Korea. The host species were identified by mitochondrial DNA (mtDNA) cytochrome oxidase I (COI) gene sequencing described by Hsu et al. (2009). The collected nematodes were identified by PCR-RFLP with subsequent sequencing and mtDNA cox2 gene sequencing, to provide information on the epidemiology of Anisakis infection in Korea.
Methods
Fresh largehead hairtail (n = 9) caught by lines were purchased from local fisheries market located in Hallim, Jeju Island, off the southern coast of the Korean Peninsula, 2015. The freshly caught fish were placed on flake ice and immediately transported to the laboratory. All of the individual fish were measured and weighed. Then, they were dissected to collect nematodes. The body cavity was longitudinally opened, and the viscera were carefully examined for collecting nematodes. The muscles were sliced, placed, and pressed between transparent glass plates, then inspected with the naked eyes under the light. All of the collected live nematodes were washed repeatedly with 0.9 % NaCl solution and preserved individually in absolute alcohol for molecular analysis. Prevalence (the number of host infected with parasites divided by the number of hosts examined) and mean intensity (the average of parasite infection among the infected hosts) were used as quantitative descriptors of the parasite population as previously described by Bush et al. (1997).
Before genomic DNA extraction, the nematodes were washed three times with PBS, then placed in 1.5 ml Eppendorf tube containing 400 μL TNES lysis buffer and crushed with a sterile pestle. Genomic DNA was extracted using the phenol-chloroform method with slight modification (Wasko et al. 2003). The extracted DNA was resuspended with 50 μL TE buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA). DNA concentration and purity were measured using Nanodrop 1000 (Thermo Scientific, USA). The ITS region (ITS1-5.8S-ITS2) of rDNA was amplified using primers A (forward: 5′-GTCGAATTCGTAGGTGAACCTGCGGAAGGATCA-3′) and B (reverse: 5′-GCCGGATCCGAATCCTGGTTAGTTTCTTTTCCT-3′) and B (D’Amelio et al. 2000). Amplification was conducted using MyCyclerTM (BioRad, USA), with the following conditions. Denaturation at 94 °C for 5 min, then 35 cycles at 94 °C for 40 s, 54 °C for 40 s, 72 °C for 90 s, and post-amplification at 72 °C for 7 min.
Restriction enzymes Hinf I, Hha I and Rsa I were used in the RFLP analysis for the amplified PCR products (D’Amelio et al. 2000). Restriction endonuclease digestion was performed using 3 μL of PCR products, 0.5 μL of restriction enzymes, 1 μL of CutSmart® buffer (NEB, USA) and distilled water up to a final volume of 10 μL. The digestion with these restriction enzymes was performed at 37 °C for 90 min. The digested products were analyzed by electrophoresis in 1.5 % agarose gel containing ethidium bromide and photographed using Gel Logic 100 Imaging System (Biostep, Germany).
For the amplification of mtDNA cox2 gene, the primers 210 (reverse: 5′-CACCAACTCTTAAAATTATC-3′) and 211 (forward: 5′-TTTCTAGTTATATA GATTGRTTYAT-3′) were used (Nadler and Hudspeth 2000). After initial denaturation at 94 °C for 5 min, 35 cycles were run at 94 °C for 40 s, 48 °C for 40 s, and 72 °C for 60 s followed by post-amplification step at 72 °C for 7 min. PCR products were purified by AccuPrep® Gel Purification Kit (Bioneer, Korea) according to the manufacturer’s instructions. 10 ng/μL of purified PCR products were directly sequenced by ABI Prism 3730 XL DNA Analyzer (PE Applied Biosystems, USA).
The sequence data of mtDNA cox2 gene was aligned with the published sequences in GenBank database (NCBI) using Clustal W method (Thompson et al. 1994). The phylogenetic tree was constructed by MEGA version 6 for comparing the genetic relationship among other published sequences by the neighbor-joining criteria (Tamura et al. 2013). The nucleotide sequences were registered in GenBank.
Genomic DNA was extracted from the muscle tissue of largehead hairtail individuals according to the methods described by Wasko et al. (2003). PCR amplification of mtDNA COI gene was conducted with the primers and conditions described by Hsu et al. (2009). PCR products were purified and sequenced by the same protocols mentioned above. The obtained sequences were aligned with other published sequences including T. lepturus, T. japonicus, and Trichiurus sp. registered in NCBI using Clustal W method (Thompson et al. 1994). The phylogenetic tree was constructed by MEGA version 6 for comparing the genetic relationship among other published sequences of Trichiurus species by the neighbor-joining criteria (Tamura et al. 2013). The nucleotide sequences were registered in GenBank.
Results
We examined nine individual largehead hairtail (mean whole body length = 90.7 ± 5.2 cm, mean body weight = 462.0 ± 47.3 g). All the examined individual largehead hairtail fish harbored nematodes (100 %, 9/9). In total, 1259 nematodes were found and mean intensity was 139.9 larvae/fish (1259/9). Most of the nematodes were found embedded in the surfaces of viscera including mesenteries (56.2 %, 708/1259) or existed freely in the body cavity (41.5 %, 523/1259). Two percent of nematodes were found in the intestines (0.6 %, 7/1259) or stomach (1.4 %, 17/1259). Only 0.3 % (4/1259) of nematodes was found in the muscles. All the information is summarized in Table 1.
aPrevalence of infection
bMean intensity
The amplification of the rDNA region produced approximately 1 kb fragment (data not shown). PCR-RFLP analysis using HinfI, HhaI, and RsaI restriction enzymes revealed different banding patterns depending on the nematodes species. Digestion of the PCR product with HinfI produced three different patterns, i.e., 350–300–250 bp, 700–350 bp, and 620–350–300–250 bp, respectively (Fig. 2a). Digestion using HhaI produced two different patterns, i.e., 550–430 bp and 350–250–200–150 bp (Fig. 2b), and digestion using RsaI produced two different patterns, i.e., 550–300 bp and 650–220 bp, respectively (Fig. 2c). These banding patterns corresponded to the known patterns of A. pegreffii (lane 1, 2 in Fig. 1a–c), Hysterothylacium sp. (lane 3, 4 in Fig. 1a–c), and hybrid genotype (A. simplex × A. pegreffii) (lane 5, 6 in Fig. 1a–c), respectively.
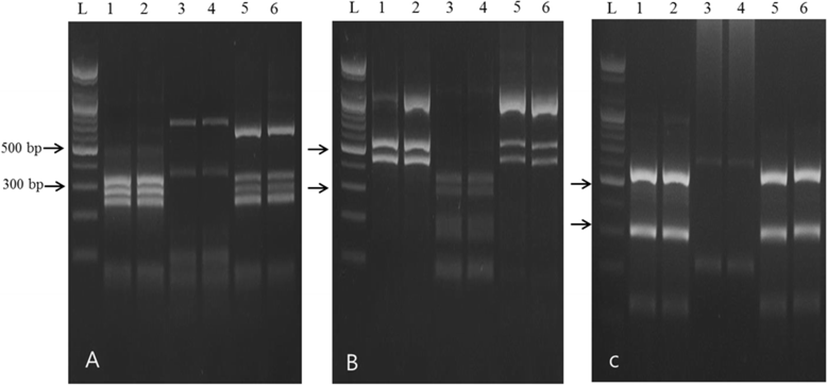
Of 1259 nematodes, most of the nematodes (98.7 %, 1243/1259) were identified as A. pegreffii. The rest of them were identified as hybrid genotype, occupying 0.5 % (6/1259) and Hysterothylacium sp., occupying 0.2 % (2/1259) of the nematodes. PCR amplification was failed for eight individuals (0.6 %, 8/1259), probably due to the failure of DNA extraction. The results are summarized in Table 2. For A. pegreffii, 20 individuals were randomly selected and sequenced for mtDNA cox2 gene because a huge number of samples were collected. For other species, all the individuals were sequenced. All of the samples generated 629 bp size products (data not shown). When the phylogenetic tree was constructed with the mtDNA cox2 sequences of these samples and those previously registered in NCBI, taxonomic position of the Anisakis species identified in this study corresponded with the results obtained by PCR-RFLP; The mtDNA cox2 sequences of A. pegreffii (Genbank accession number: KU921231-921250) were clustered with those isolated from chub mackerel in Korea and Japan (Fig. 2), with 97.9–100.0 % homology. The mtDNA cox2 sequences of Hysterothylacium sp. showed the highest homology (92.0 %) with Hysterothylacium sp. isolated from chub mackerel (Genbank accession number: KC633443). The mtDNA cox2 sequences of all the hybrid genotype specimens were found to be A. pegreffii (data not shown).
Microhabitat/species name | A. pegreffii | Hybrid | Hysterothylacium sp. | Fail | Total | |
|---|---|---|---|---|---|---|
Viscera | Intestines | 7 | 0 | 0 | 0 | 7 |
Stomach | 17 | 0 | 0 | 0 | 17 | |
Mesenteries | 701 | 3 | 0 | 4 | 708 | |
Body cavity | 514 | 3 | 2 | 4 | 523 | |
Muscle | 4 | 0 | 0 | 0 | 4 | |
Total | 1243 | 6 | 2 | 8 | 1259 | |

PCR amplification and sequencing of mtDNA COI sequences from fish samples revealed that all of the nine samples produced one single band of 630 bp (data not shown). The obtained sequences (Genbank accession number: KU963588, 963589) were clustered with those of T. japonicus previously registered in GenBank, with 98.4–99.8 % homology (data not shown).
Discussion
Human infection with A. simplex L3 accidentally occur when raw or undercooked fish contaminated with the L3 is ingested. Although live L3 can rarely develop to L4 or adults in the human gastrointestinal tracts, they can cause mild to severe epigastric pain, nausea, vomiting, and diarrhea (Audicana and Kennedy 2008). In addition, allergic reactions occur in some patients, which may occur even by the ingestion of dead A. simplex larvae (Audicana and Kennedy 2008). In fish, the larvae are mainly located freely in the visceral cavity or encapsulated as flat, tight spirals in and on the visceral organs; however, a minor proportion may migrate into the muscles, which may draw attention of consumers (Levsen and Berland 2012). Thus, it is necessary to have knowledge about the host range of each Anisakis species, together with the clear identification and their geographical distribution to avoid accidental human infection and warrant food safety issue.
In this study, most of the nematodes were found throughout the viscera (56.2 %) or from the body cavity of the largehead hairtail (41.5 %) (Table 1). Those from the viscera were embedded in or stick to the surfaces of the mesenteries and intestines, whereas those from the body cavity were freely moving in the body cavity. On the other hand, only 0.4 % (6/1,420) of the nematodes was found in the muscles.
Of 1259 nematodes, most of the nematodes (98.7 %, 1243/1259) were identified as A. pegreffii by PCR-RFLP and direct sequencing. Other species, Hysterothylacium sp. and hybrid genotype (A. simplex × A. pegreffii) were rarely found.
A. pegreffii Campana-Rouget and Biocca, 1955 is the dominant species of Anisakis in the Mediterranean Sea and also widely distributed in the austral region between 30° N and 55° S. Many pelagic and demersal fish species are known to be the intermediate/paratenic hosts caught in this area, and toothed whales (Delphinidae, Ziiphidae, Physeteridae) and baleen whales (Neobalaenidae) are known to be the definitive hosts (Mattiucci and Nascetti 2008). A. pegreffii has been also reported frequently at the larval stage in many fish species in East Asian waters (Bak et al. 2014 and the references therein). Recent reports in Italy suggest that A. pegreffii can be the etiological agent of human infection (Mattiucci et al. 2013).
Genus Hysterothylacium Ward and Magath, 1917 is one of the common genera in the family Raphidascarididae (Fageholm 1991), in which there are approximately 70 species (Shamsi et al. 2013). The adult stages are usually found in the alimentary canal of predatory teleost, while the L3 are found encapsulated in the mesentery and viscera of various species of marine teleosts and also in various marine invertebrates (Keskin et al. 2015). Thus, it is not host specific in the larval stages, exhibiting a global distribution (Shamsi et al. 2013). Many species of the genus Hysterothylacium have been poorly described; thus, their genetic characteristics have been frequently deposited in Genbank without clear identification at the species level. Although the RFLP patterns in this study was in accordance with those of Hysterothylacium sp. described in Bak et al. (2014), we could not identify these specimens at the species level because the highest homology of the mtDNA cox2 sequences showed 92 % with that of Hysterothylacium sp. registered in GenBank. It would be necessary to conduct morphological observation and further molecular analysis, for clear identification of these specimens.
The prevalence of infection and mean intensity for anisakid nematodes L3 in T. japonicus calculated in this study were extremely higher than those described by Lee et al. (2009). There can be several explanations for this difference. In our study, we collected freshly caught T. japonicus from Korean waters, whereas they collected T. lepturus in a fisheries market, without information on their geographical origin.
In marine food webs, the abundance of phytoplankton is strongly related to temperature and changes in phytoplankton biomass can affect all subsequent stages, first impacting on marine crustaceans, the predominant group in zooplanktons abundance and biomass, and thereafter migratory fish, cephalopods, and predatory marine mammals further up the food chain (Utaaker and Robertson 2015). The life cycles of Anisakis species involve various hosts at different levels in marine food webs. The high prevalence of Anisakis species in fish hosts in this study is therefore thought to reflect the richness of micro/macroinvertebrates and possibly subsequently have relations with the biomass of other predators such as pinnipeds and cetaceans in waters investigated. Several papers also reported relatively a high prevalence of anisakid nematodes larvae in the largehead hairtail (Borges et al. 2012; Kong et al. 2015; Umehara et al. 2010), as in this study. The low number of the nematodes larvae in the previous study (Lee et al. 2009) may just reflect the low abundance of micro/macroinvertebrates in the areas where the largehead hairtail were caught.
Several papers described anisakid nematodes L3 from the largehead hairtail (Borges et al. 2012; Kong et al. 2015; Lee et al. 2009; Shih 2004; Umehara et al. 2010; Zhang et al., 2013), but there was no report in which T. japonicus was mentioned as host species, except for our study. It is not clear if the host species mentioned was in fact T. japonicus or other Trichiurus species in their studies because the identification of host species was not clearly conducted. But except for the paper by Borges et al. (2012), they probably sampled T. japonicus because the known geographical distribution of T. japonicus, the northwestern Pacific (Hsu et al. 2009; Tzeng et al. 2007) includes their sampling areas.
After Anisakis L3 in the invertebrate or primary fish hosts are ingested by predatory fish, they penetrate in the intestinal wall and then may remain within the body cavity or migrate into the musculature or internal organs of the fish. The different relative abundance of these L3 among these microhabitats in fish can be affected by many factors such as the parasites species, the fish species, the fish age, and the environmental conditions (Lymbery and Cheah 2007). Although the mechanisms of larval penetration within fish hosts are still unknown, some studies revealed that L3 migrate from the visceral organs to the muscle after the death of the fish hosts and this postmortem migration is thought to be also affected by interactions among parasites, host, and external environments (Cipriani et al. 2016; Lymbery and Cheah 2007). Thus, the risk of human infection can be enhanced if the fish are ingested whole or if the fish have been kept without evisceration for a period of time.
Several methods such as postharvest handling, freezing, and adequate cooking are suggested for prevention or control measures of the human infection by ingestion of raw or undercooked fish harboring Anisakis L3 (Audicana and Kennedy 2008). Deep freezing and/or adequate cooking remain the most effective measures to prevent human infection (Audicana and Kennedy 2008; Lymbery and Cheah 2007). In addition, immediate evisceration or freezing of fish after catch will be necessary for the prevention of human infection (Lymbery and Cheah 2007). In case of our study, most of the Anisakis L3 on the largehead hairtail was found from the body cavity and viscera, which are generally not consumed raw and removed before cooking. In addition, A. simplex (s.s.), the most commonly involved species in human infection, was not found in this study. Thus, the largehead hairtail may not be the common source of human infection in Korea. But care should be still taken because A. pegreffii is also considered as a possible human pathogen (Mattiucci et al. 2013), and a high prevalence of anisakid nematode larvae in the largehead hairtail may raise food safety problems to the consumers. Immediate evisceration or freezing of fish after catch will be necessary before consumption.
Conclusions
Information on the anisakid nematodes fauna of commercially important fish species is important because the L3 of anisakid nematodes, mostly A. simplex, can cause accidental human infection in countries where the customs of eating raw or undercooked fish or cephalopods exist. Given the growing popularity of these customs worldwide, the risk of human infection is widespread, together with the increased number of new host records and expansion of the geographical ranges of Anisakis spp. The largehead hairtail, one of the commercially important fish species in Korea, was investigated for anisakid nematodes fauna. Most of the nematodes were found throughout the viscera (58.1 %) and the body cavity (41.5 %), which is not consumed raw. Only 0.3 % was found in the muscles. A. pegreffii was the most frequently encountered nematode species (98.7 %) from the largehead hairtail in this study. Detailed information on the distribution of different Anisakis species in the largehead hairtail in this study has significant implications for the biology, ecology, and epidemiology of anisakid nematodes.








