Background
Antifreeze proteins (AFPs) lower the freezing point but not the melting point of aqueous solutions by inhibiting the growth of ice crystals via an adsorption-inhibition mechanism (DeVries and Wohlschlag 1969; Raymond and DeVries 1977; Jia and Davies 2002). Binding of AFP to the surface of an ice crystal inhibits further growth of the crystal, which creates a temperature gap between the melting and freezing points of the aqueous solution. This gap, called thermal hysteresis (TH), is used to quantitatively express the activity of AFP. AFPs are essential for polar fish to survive at the subfreezing temperature of seawater (DeVries and Wohlschlag 1969; DeVries 1971). The freezing point of fish blood can be lowered down to −0.7 °C by solutes in the serum, and an additional 1 to 1.2 °C depression is achieved by AFPs. The total freezing point depression of approximately 1.9 °C allows the fish blood to remain unfrozen even at −1.9 °C, the temperature of polar seawater during winter (DeVries 1971; Davies and Hew 1990; Raymond and DeVries 1977; Fletcher et al. 2001). To date, four fish AFPs with distinct TH activities have been identified: antifreeze glycoproteins (AFGPs) and AFP types I, II, and III (Davies and Hew 1990). Their TH activities are typically approximately 1 °C, which is sufficient to protect the fish from freezing.
Recently, AFPs and antifreeze activity have also been reported from fish living in regions other than polar regions such as north-temperate and temperate areas (Deng et al 1997; Gauthier et al. 2008; Nishimiya et al. 2008; Kim 2015). Conventionally, north-temperate fish off the east coast (50° N) of Canada and/or Hokkaido (43.06° N) have been suggested to possess AFPs as a result of convergent evolution (Cheng 1998; Cheng and Chen 1999; Cheng 1998; Cheng et al. 2003; Graham et al. 2008, 2013), as sea ice forms in these coastal areas during winter similar to that in the austral winter at the Antarctic. Conversely, it is not likely that temperate and/or subtropical fish require antifreeze activity for survival. However, contrary to our expectation, we observed antifreeze activity in some temperate and subtropical fish from the ice-free East Sea of Korea (Kim 2015). Among nine fish examined in that region, three fish, Gymnocanthus herzensteini, Zoarces gillii, and Kareius bicoloratus, exhibited antifreeze activity. Furthermore, during the investigation, tissues of Paralichthys olivaceus did not display discernible antifreeze activity although analysis of its expressed sequence tags (ESTs) revealed an EST encoding a putative type IV antifreeze protein (AFP IV) from a liver complementary DNA (cDNA) library.
AFP IV was first identified in the longhorn sculpin, Myoxocephalus octodecemspinosus, which inhabits the north-temperate coastal waters of North America (Deng et al. 1997; Deng and Laursen 1998). However, unlike other types of AFPs, the function of AFP IV remains questionable. This uncertainty arises as the AFP IV THs from longhorn and shorthorn sculpins are on the verge of detectable limits, the physiological concentration of this protein in adult fish blood is too low to function as a biological antifreeze (Deng and Laursen 1998; Zhao et al. 1998; Gauthier et al. 2008), and because its homologues are present in many fish from all the oceans ranging from tropic to polar as well as in freshwater fish, many of which do not require an antifreeze protein. In addition, AFP IV shares 20 % sequence identity with the apolipoprotein superfamily (Cheng 1998; Gauthier et al. 2008). Based on the initial characterization of two AFP IVs from longhorn and shorthorn sculpins, Gauthier et al. (2008) speculated that AFP IV may not have been selected and evolved as a main antifreeze owing to the presence of the more efficient type I AFP. In addition, very recently, a Chinese group reported that two AFP IV genes, afp4a and afp4b, from the tropical fish gibel carp (Carassius auratus gibelio) and zebrafish (Danio rerio) are essential for epiboly progression and convergent movement during zebrafish embryo gastrulation (Liu et al. 2009; Lee et al. 2011; Xiao et al. 2014). This evidence is consistent with previous findings that some apolipoproteins, which are homologues of AFP IV, play a significant role in embryonic morphogenesis and organogenesis in these fish (Xia et al. 2008; Choudhury et al. 2009; Zhang et al. 2011; Wang et al. 2013; Xiao et al. 2014). Together, these results imply that the main function of AFP IV is not as an antifreeze and that its gain of ice-binding capacity might not be the consequence of selective pressure of a cold environment but may have arisen by chance. If the ice-binding affinity of AFP IV found in polar and other northern fish had evolved simply by selective pressure from the cold environment, then its homologues in temperate and tropical fish would not be expected to possess a similar affinity. To address this issue, we cloned and analyzed cDNA of the subtropical fish P. olivaceus, expressed its recombinant protein in Escherichia coli, and measured its TH activity.
Methods
An adult olive flounder cultured in Korea was purchased from a local fish market, and cDNA libraries were constructed for the liver and muscle tissues. Briefly, the tissues were ground using a mortar and pestle in liquid nitrogen. mRNA was isolated from the ground liver and muscle tissues using the Micro-FastTract™ 2.0 Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Unidirectional cloning of cDNA was performed using a ZAP-cDNA synthesis kit (Stratagene, Santa Clara, CA, USA) as described in the user manual. Mass phagemid vector excision was performed, and the mixture was plated on a Luria-Bertani (LB) agar plate. The excised colonies were randomly selected and incubated separately in 2 mL LB broth with ampicillin (50 μg/mL). The cultured cells were harvested by centrifugation, and the plasmids were isolated using a standard phenol method. The plasmids were then digested with XhoI and EcoRI and electrophoresed on a 1.5 % agarose gel. The plasmids containing the 1-kb insert were selected and sequenced with the SK primer (5′-CGC TCT AGA ACT AGT GGA TC-3′) for 5′-directional analysis. The trimming of vector and linker sequences from the raw sequences was performed manually using Chromas software (http://technelysium.com.au/wp/chromas.html). NCBI databases were then searched for sequences homologous to each EST using BlastN and BlastX.
To obtain 5′ end sequences of the afp4 gene, the CapFishing™ full-length cDNA Premix Kit (Seegene, Korea) was used. For 5′-rapid amplification of cDNA ends (RACE), the target primer R1 (5′-CAA GCC GCT CCA TGA ACC T-3′) was designed based on the 3′-untranslated region (UTR) of the afp4 gene. The polymerase chain reaction (PCR) product was isolated, cloned using the TOPO TA Cloning Kit (Invitrogen), and sequenced.
For genome structure analysis, specific forward (5′-ATG AAA TTC TCC CTC ATT GC-3′) and reverse (5′-GGC TTG TTT AGT TGG AGA TG-3′) primers were designed based on the afp4 cDNA sequence. The amplified PCR product of 1.2 kb was sequenced and used as a probe to screen for bacterial artificial chromosome (BAC) clones containing the target genomic region. A BAC library of olive flounder (Dr. Aoki, Tokyo University of Fisheries) was arrayed on nylon membranes in a 3 × 3 grid pattern using a Biomek 2000 Workstation (Beckman Coulter, Brea, CA, USA) and hybridized with the 32P-labeled PCR product. The plasmids of six positive clones (A1–A6) were isolated using the phenol method. The plasmid of A3 was digested with EcoRI, KpnI, BamHI, PstI, or HindIII, and each digested DNA was electrophoresed on a 0.7 % agarose gel and transferred to a positively charged nylon membrane. The membrane was then hybridized with the 1.2-kb digoxigenin-labeled probe previously amplified by PCR (Roche, Roswell, GA, USA). The plasmids of A2–A6, except for A5, were full-digested with EcoRI and electrophoresed on a 0.7 % agarose gel, transferred to a nylon membrane, and also hybridized with the above probe. The 2.9-kb fragment of the A3 plasmid digested with EcoRI was eluted from the agarose gel and ligated into an EcoRI-digested pUC19 vector. The plasmids were then transformed into bacteria, isolated, and sequenced.
To construct the expression vector harboring the flounder afp4 gene, the mature coding region was amplified from the cDNA library using the forward primer, 5′-CAT ATG CAA GAT GCT GCT GAT CTG-3′, including an NdeI site and a reverse primer, 5′-TCT AGA TTA GTT GGA GAT GCT GCG-3′, including an XbaI site using Pfu polymerase. After A-tailing using Taq polymerase, the PCR product was cloned into the TOPO TA cloning vector (Invitrogen). The plasmids were isolated and confirmed by sequencing. The insert DNAs digested with NdeI and XbaI were subsequently ligated into pCold I expression vectors (TaKaRa, Otsu, Japan), digested with the same restriction enzymes. The resulting plasmid was transformed into E. coli BL21.
For protein expression, the E. coli transformants were inoculated in LB medium with 50 μg/mL ampicillin and grown at 37 °C overnight. The seed culture was diluted 100 times in the LB medium with 50 μg/mL ampicillin and grown at 37 °C until OD600 reached 0.5, at which time the culture was transferred to a 15 °C shaker and further incubated for 1 h. Then, isopropyl β-d-thiogalactopyranoside (IPTG) was added to the culture medium at a final concentration of 0.5 mM to induce the AFP IV protein. The cells were incubated further for 15–20 h at 15 °C in a shaker. The recombinant AFP IV was subsequently purified from the soluble supernatant using Ni-affinity chromatography. The AFP IV-overexpressed cell pellet was suspended in 20 mM Kpi, pH 7.4, 50 mM NaCl, and 5 mM imidazole and lysed using sonication. The supernatant of the cell lysate was loaded onto a Ni-NTA column pre-equilibrated with a lysate buffer. The AFP IV was eluted with a buffer containing 20 mM Kpi, pH 7.4, 50 mM NaCl, and 500 mM imidazole. The fractions containing the AFP IV protein were pooled and dialyzed against a lysis buffer and concentrated using ultrafiltration. The molecular mass of the recombinant AFP was determined using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) using sinapinic acid (3,5-dimethoxy-4-hydroxy cinnamic acids) as a matrix with the Voyager-DE STR Biospectrometry Workstation (Applied Biosystems, Foster City, CA, USA).
TH activity and ice crystal morphology were examined using a nanoliter osmometer (Otago Osmometers, Dunedin, New Zealand) connected to a cold well stage mounted on a light microscope equipped with a Canon digital camera (Tokyo, Japan). A droplet containing a few nanoliters of the sample to be assayed was layered into a well filled with oil. The sample well was placed on the stage and frozen rapidly at approximately −20 °C. The temperature was raised slowly until a single ice crystal remained. Then, the temperature was lowered again slowly while the ice crystal morphology was maintained. The ice crystal image was pictured at ×40 magnification. The melting point of the sample was taken as the temperature at which a single ice crystal formed, and the freezing point was the temperature at which rapid growth of the ice crystal was observed. The difference between melting and freezing points is considered the TH value. While measuring the TH values, changes of ice shape were also observed.
Results
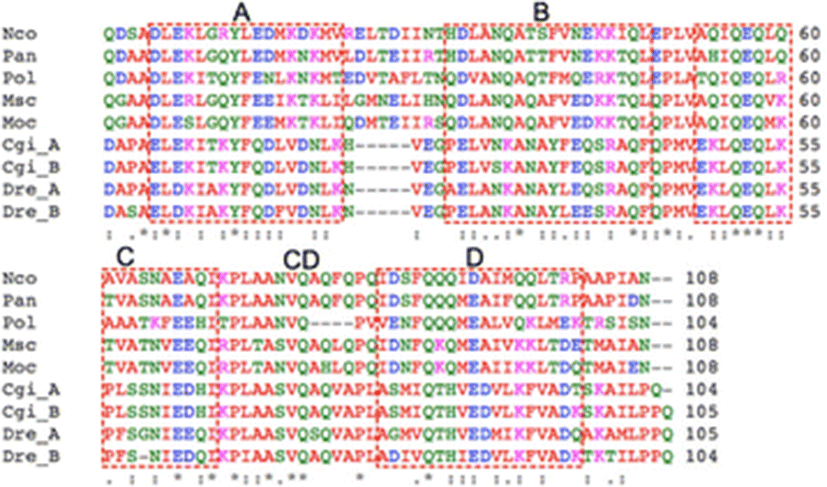
A total of 149 plasmids that contained relatively large inserts were isolated and sequenced from liver and muscle cDNA libraries of P. olivaceus (data not shown). Of these, one of the liver ESTs displayed high similarity to the afp4 gene of longhorn sculpin (M. octodecimspinosus) (Deng et al. 1997; Deng and Laursen 1998; Zhao et al. 1998). The full-length cDNA was obtained using RACE-PCR. The nucleotide sequence of the full-length cDNA consisted of 51 nt 5′-UTR, 375 nt open reading frame (ORF), and 174 nt 3′-UTR. The polyadenylation signal (ATTAAA) was found between 15 and 19 nt upstream of the polyadenylation site. The ORF encodes a protein of 124 amino acid residues. The SignalP 3.0 program (Bendtsen et al. 2004) predicted the first 20 residues in the N-terminus as a signal peptide for secretion. As shown in Fig. 1, mature AFP IV amino acid sequence alignment demonstrated high identities with functional AFP IVs from polar fish, 55 % identity with Pleuragramma antarcticum, 53 % with M. octodecemspinosus, 52 % with Myoxocephalus scorpius, and 49 % with Notothenia coriiceps. However, it showed relatively lower identities (less than 30 %) to AFP IVs from tropical fish such as zebrafish (D. rerio) and gibel carp (C. gibelio). The calculated molecular masses of the precursor and the mature P. olivaceus AFP IV were 13,945 and 11,941 Da, respectively.
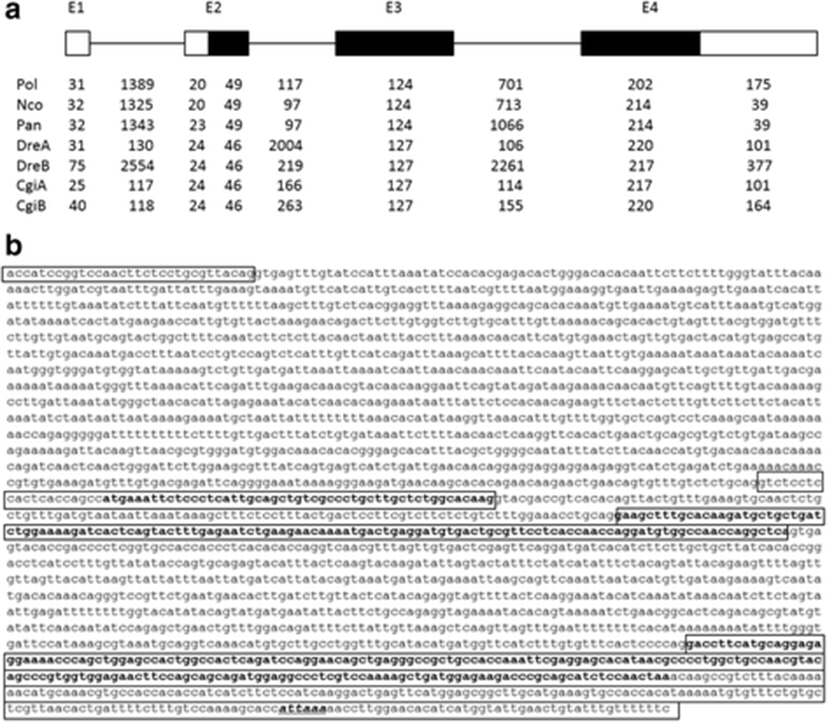
We identified six positive BAC clones, A1–A6, by hybridization screening, all of which except for the A5 clone contained the afp4 gene, which was further confirmed by Southern blotting (data not shown). To obtain the genomic sequences, an approximately 2.9-kb genomic fragment of A3 was subcloned and sequenced. However, this fragment did not contain about 740 bp after the first exon; we therefore amplified the gap region by PCR using specific primers based on the partial genome sequence. We determined that the genomic structure of this gene consisted of four exons and three introns (Fig. 2a) comprising a total of 2808 bp. The intron/exon boundaries were well conserved according to the GT-AG rule. Based on the cDNA and genomic sequences, the afp4 gene contains three translated exons (exons 2–4) and one non-translated exon (exon 1). The length of intron 1 is 1389 bp. Intron 2 (117 bp) is significantly shorter than the other introns. Exon 1 and 20 nt of exon 2 encodes the 5′-UTR. In exon 2, the ATG translation initiation site is located 21 bp from the 5′ end of exon 2. Exons 2, 3, and 4 encode 16, 41, and 67 amino acids, respectively.
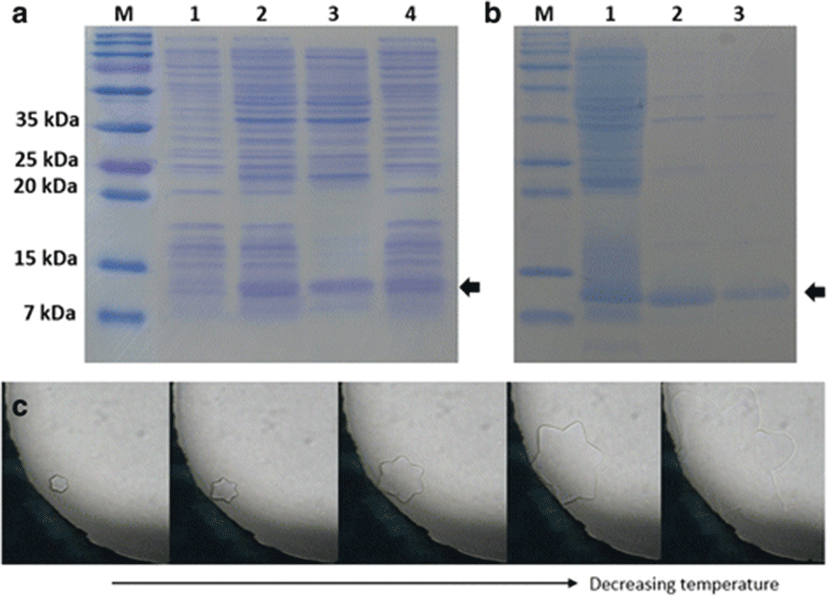
The mature AFP of olive flounder was expressed in the pCold I vector with an additional N-terminal peptide consisting of a translation-enhancing element, factor X cleavage site, and 6 His-tag. Although the recombinant AFP was expressed as a mixture of soluble and insoluble forms, soluble protein was successfully purified to relative homogeneity using His-tag affinity chromatography (Fig. 3a, b). Although as shown in Fig. 3b there were a few contaminants visible in the eluted fractions and concentrated solution, because there is no intrinsic antifreeze activity from E. coli and any antifreeze activity observed was solely from the recombinant AFP IV, we did not attempt to remove the contaminants completely. The calculated molecular mass of AFP with the additional N-terminal sequence was 14,097 Da. The purified protein migrated faster by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE); however, its molecular mass was confirmed by MALDI-TOF. The TH activity and ice crystal morphology of AFP were detected for both the soluble fraction of the cell lysate and the purified protein. TH activity of the cell lysate was approximately 0.04 ± 0.01 °C, which indicated that the soluble fraction contained active AFP IV. The AFP IV was purified but concentrated only up to 0.5 mg/mL using ultrafiltration because it tended to aggregate at higher concentrations of greater than 0.5 mg/mL. The TH value was 0.07 ± 0.01 °C at the concentration of 0.5 mg/mL. As the purified AFP IV was more concentrated than the soluble cell lysate, the TH value was higher. Similar to other AFP IVs, the olive flounder AFP produced a star-shaped ice morphology (Fig. 3c).
Discussion
The reason underlying the presence of a gene for AFP IV in P. olivaceus (Pol afp4) is unclear, as it is a subtropical fish that does not require AFP for survival. In addition, P. olivaceus did not initially demonstrate an antifreeze activity (Kim 2015). However, its EST analysis revealed that it carried a transcribable afp4 gene, which further raised the question of the function of this transcript. To test the hypothesis that the ice-binding ability of AFP IV was gained specifically by the selective pressure of cold environments, we cloned the Pol afp4 gene and evaluated its TH activity.
Previously, Cheng (2003) proposed that the longhorn sculpin AFP IV had evolved from a domain duplication of apolipoproteins through the selection pressure of a subfreezing marine environment. On the other hand, its homologues have been identified in many fish isolated from polar to tropic seawater as well as from freshwater, and their gene organization is also very similar (Gauthier et al. 2008; Lee et al. 2011; Xiao et al. 2014). A comparison of the genome structures of known fish AFP IV proteins including the P. olivaceus afp4 gene characterized in the current study showed that the exon and intron positions are highly conserved (Fig. 2a), supporting the suggestion that fish afp4 genes resembled each other regardless of their habitats. However, the sequences and sizes of the introns differ among fish. Notably, the genomic organization of the P. olivaceus afp4 gene exhibits greater similarity to those of AFP IV from two Antarctic fish, P. antarcticum and N. coriiceps (Lee et al. 2011), than to tropical carp and zebrafish. In particular, the two afp4 genes found in both carp and zebrafish are arranged in a head-to-tail tandem manner. This tandem arrangement is believed to be due to gene duplication (Xiao et al. 2014). However, only a single afp4 gene is present in the genomes of two Antarctic fish and P. olivaceus, with no evidence of gene duplication being evident. Together, this evidence renders the establishment of the evolution of this gene more challenging.
The sequence alignment showed that the four additional AFP IVs with a known antifreeze activity share a 48–53 % amino acid sequence identity with Pol AFP IV (Fig. 1). Deng and Laursen (1998) proposed a four-helix bundle model for AFP IV based on the high sequence identity to an apolipoprotein. Notably, as shown in Fig. 1, Pol AFP IV carries a four-residue deletion that can be mapped onto the linker between C and D helices (CD linker) linker of the four-helix bundle model (Gauthier et al. 2008; Lee et al. 2011). However, despite the deletion in the linker, Pol AFP IV exhibits almost the same TH activity as other AFP IV proteins (Deng et al. 1997; Deng and Laursen 1998; Zhao et al. 1998; Gauthier et al. 2008; Lee et al. 2011), which strongly indicates that residues important for ice-binding are conserved in Pol AFP IV and not located in the CD linker. This result also implies that the deletion is not likely to disrupt the four-helix bundle structure, because the characteristics of recombinant Pol AFP IV are similar to those of other recombinant AFP IVs. This may hold positive implications for AFP IVs from fresh- and warmwater fish as well (Xiao et al. 2014). Thus, even though the AFP IVs from C. gibelio and D. rerio each contain a deletion in the loop connecting helices A and B (Xiao et al. 2014), they might therefore have very similar three-dimensional folds as there are no drastic changes in the sequences corresponding to the four helices.
In particular, although very weak compared to AFPs I, II, and III at the same concentrations, recombinant Pol AFP IV exhibits TH activity of 0.07 ± 0.01 °C at 0.5 mg/mL. This activity is comparable to those of the Antarctic AFP IVs (Lee et al. 2011) but is slightly lower than those of recombinant shorthorn and longhorn sculpin counterparts (Deng et al. 1997; Deng and Laursen 1998; Gauthier et al. 2008; Zhao et al. 1998). Although the natural AFP IV of longhorn sculpin begins to aggregate at concentrations above 25 mg/mL (Deng et al. 1997), recombinant AFP IVs expressed in E. coli were less soluble and prone to aggregate at concentrations as low as 0.4 mg/mL for M. octodecemspinosus AFP IV, 0.5 mg/mL for P. antarcticum and N. coriiceps AFP IVs, and 10 mg/mL for M. scorpius AFP IV (Gauthier et al. 2008; Lee et al. 2011). Similar to these AFPs, Pol AFP IV was also only able to be concentrated up to 0.5 mg/mL. However, this low concentration is comparable to the physiological concentration (0.05–0.1 mg/mL) of AFP IV in the blood plasmas of shorthorn sculpin as detected by immunoblotting (Gauthier et al. 2008). At this low concentration, the TH value of the natural longhorn sculpin AFP IV is similar to those of other recombinant AFP IVs (Deng et al. 1997; Deng and Laursen 1998), suggesting that if the recombinant proteins were able to be concentrated, their TH values would be quite close to that of the longhorn sculpin protein. The TH values from AFP IVs are not enough to keep fish from freezing at subzero temperature, addressing the question of their real function in vivo.
The ice morphology shaped by Pol AFP IV was also similar to those produced by the Antarctic fish and shorthorn sculpin AFP IVs (Gauthier et al. 2008; Lee et al. 2011). The star-shaped ice crystals represent weak antifreeze activity, which is also consistent with the low TH values. Notably, if longhorn sculpin AFP IV had been evolved only through the selective pressure of the cold Antarctic environment approximately 10–15 million years ago, then there would likely be a very slim chance of comparable AFP IV evolution in temperate, subtropical, and/or tropical fish such as P. olivaceus, which might have experienced very recent temporal glaciation approximately 2.5 to 3 million years ago. Instead, the abundant expression of AFP IVs in carp and zebra fish observed in oocytes and embryos strongly support its association with early development (Goetz et al. 2006; Liu et al. 2009; Brenton et al. 2012).








