Background
Concerns have risen over the presence of antimicrobial residues in food fishes because the contaminants can cause human pathogenic bacteria to develop resistance to therapeutically valuable antimicrobials (Cabello, 2006). Development of antibacterial resistance will then require higher doses of the same agent, or more potent drugs for efficacious treatment of bacterial pathogens. Of several known mechanisms for resistance development, degradation of antimicrobial agents is regarded as the most frequent one (De Pascale and Wright 2010).
Soon after the introduction of β-lactam antibiotics such as penicillins and cephalaosporins into clinical use, bacterial resistance to these drugs were reported (Abraham and Chain, 1988). Those bacteria have been found to harbor plasmid-borne genes producing enzymes which can catalyze a few β-lactam antibiotics nullifying the efficacy of such drugs (Matagne et al. 1999). However, the efficacy has become increasingly limited by the advent of β-lactamase that catalyzes a broader range of β-lactam antibiotics compared with previous enzymes (Livermore 1995). Those enzymes are called extended-spectrum β-lactamase (ESBL) and they are usually represented by TEM-52.
If β-lactamase can catalyze β-lactam antibiotics in fish tissues, it could be useful to remove residual antibiotics remaining after use to food fish species. To test this possibility, we assessed the catalytic activity in vitro of purified TEM-52 enzymes obtained by recombinant DNA techniques, followed by an in vivo activity test in carp after administration of cefazolin, a β-lactam antibiotic.
Methods
The gene for TEM-52 was obtained from a Korean clinical isolate after necessary cloning processes (Pai et al., 1999). Its sequence was confirmed to match 100% by query coverage tests with those of extended-spectrum enzymes reported from Salmonella enterica subspp. enterica serovar Typhimurium (gb|AY883411.1|) and Klebsiella pneumoniae (emb|Y13612.1). This gene was used to build a recombinant plasmid and inserted into E. coli and then cultured at 30 °C for about 48 h in 2-L complex media. When the culture reached the density of ca. 0.7 optical density at 650 nm, cells were harvested by centrifugation (×13,000g, 10 min, 3 °C). Cells were disrupted by sonication and the lysate was processed for purification purposes with Q-Sepharose FF column chromatography after refolding steps. The refolding process was composed of decreasingly stepwise dilutions at 8, 2, 1, and 0.4 M in denaturant urea. The TEM-52 enzyme was eluted in 20 mM Tris buffer (pH 7.0) and adjusted to contain 500 mU/ml β-lactamase activity when assayed for 5 min using nitrocefin (50 μg/ml, Oxoid, Basingstoke, UK) as the substrate (O’Callaghan et al., 1972). The final purified TEM-52 enzyme is known to have a molecular mass of 28 KDa (Perilli et al., 2002).
Degradation activity of TEM-52 against various classes of antimicrobials was assessed incubating the β-lactamase enzyme solution (25 mU/ml) with various antimicrobials. Ten different antimicrobials (cefotaxime-Na, cefazolin-Na, penicillin-G-K salt, amoxicillin, pivampicillin, oxytetracycline-HCl, ciprofloxacin, erythromycin, josamycin, and streptomycin) were incubated with TEM-52 in 50 mM sodium phosphate buffer (pH 7.0) for 30 min at 25 °C, and then followed by bioassay for semi-quantitative analysis of remaining drug levels. These antimicrobials were obtained from Sigma (St. Louis, MO, USA) except pivampicillin (Dong-Hwa Pharmaceuticals, Seoul, Korea) and josamycin (Wako Chemicals, Japan). Paper disks (8 mm diameter, Advantec Toyo, Dublin, CA, USA) were soaked with 50 μl of incubated enzyme-antimicrobial mixtures and placed on top of BHI agar in Petri dishes streaked with Staphylococcus epidermidis ATCC10145 (precultured in BHI to 1.8 × 107 CFU/ml). The dishes were incubated at 36 °C for 12 h, and inhibitory zones beyond the disk areas were measured.
Of several β-lactam antibiotics, cefazolin was selected as the model antibiotic because of the simplicity in chemical analysis for both in vivo and ex vivo studies. To assess degradation activity of TEM-52 more quantitatively than the above in vitro study, cefazolin was incubated with TEM-52 (25 mU/ml in 50 mM phosphate buffer, pH 7.0) for 30 min and remaining concentration of cefazolin was chemically analyzed with a high-performance liquid chromatography (HPLC) method.
The HPLC-UV method for cefazolin analysis (Nadai et al., 1993) was composed of a Waters 2690 separation module and Waters 2487 dual wavelength UV-visible detector at 274 nm (Waters, Milford, MA, USA). Samples were injected into a C18 reverse-phase HPLC column (250 × 4.6 mm, 5 μm particle size, Shiseido, Japan) at 10–50 μl range and eluted with a mobile phase composed of 30 mM sodium phosphate buffer (pH 5.0) and methanol (88:12 ratio). Flow rate of the mobile phase was 1.2 ml/min.
For in vivo cefazolin degradation experiment, common carp Cyprinus carpio weighing 40–100 g maintained at 23 °C were used. Cefazolin (sodium salt, Sigma) was dissolved in sterile saline and administered intramuscularly (im) around the lateral line to render doses of 10 and 30 mg/kg of drug base. TEM-52 enzyme was immediately injected into the peritoneal cavity at 75 mU/100 g body weight. This intraperitoneal (ip) injection volume was 150 μl/100 g and an equivalent volume of saline was also administered to control fish. One hour after TEM-52 injections, fish were anesthetized with MS-222 (Sigma) and blood was sampled for cefazolin analysis through the caudal vessels. Serum was subsequently obtained following centrifugation at ×3600g for 20 min under refrigeration (3 °C). Liver and muscle were also isolated for cefazolin analyses. Serum and fish tissues were mixed with HPLC mobile phase in tenfold volumes and vigorously vortex-shaken before filtering through membrane filters, and finally injected into the HPLC column. This method led to an almost complete cefazolin recovery (>90%, n = 3). With cefazolin-spiked liver and muscle tissues (10, 20, 50, 100, and 200 ng/g), linearity of the standard curve was confirmed (r2 = 0.967). The limits of detection (LOD) and quantification (LOQ) were 10 and 30 ng/g, respectively.
In some fish, TEM-52 was administered into the peritoneal cavity (ip) at 75 mU/100 g to naive carp, and serum was obtained 1 h later to assess ex vivo degradation activity against cefazolin. All experiments using carp were performed in accordance with the guidelines approved by the Institutional Animal Care and Use Committee, Kunsan National University, Korea.
Results and discussion
Table 1 shows in vitro enzymatic activity of TEM-52 to degrade different classes of antibacterial agents. TEM-52 is a wide spectrum β-lactamase obtained from E. coli through recombinant DNA technique. The enzyme preparation reduced antibacterial activity of β-lactam antibiotics without effects on drugs of other classes, indicating highly selective degradation activity toward β-lactams. Other classes of antibacterials, i.e., tetracycline, quinolone, macrolide, and aminoglycoside, were not influenced at all. These data reflect that the enzyme preparation contains exclusively β-lactamase efficacy and hydrolyzes β-lactams, cephalosporins, and penicillins (Bush et al., 1995; Pai et al., 1999; Poyart et al., 1998; Shahada et al., 2010), without any activity on other antibacterials.
Concentrations of antimicrobials in incubation were adjusted to produce 6–11 mm clear zones beyond the disk areas (0.5–500 μg/disk)
Triplicate determinations
Of the five β-lactam antibiotics examined with the in vitro bioassay, cefazolin was chosen for in vivo studies mostly because the procedures for HPLC analysis are quite simple. The results obtained after simultaneous administration of TEM-52 and cefazolin to carp are demonstrated in Fig. 1. In all three tissue samples examined, the levels of cefazolin were significantly lower in TEM-52 co-administered fish compared with the cefazolin-alone group. With cefazolin doses 10 and 30 mg/kg, and regardless of tissues, the degree of reduction was ∼50% (Fig. 1a–c). In this in vivo experiment, TEM-52 was administered into the peritoneal cavity whereas cefazolin into the muscle. These results therefore indicate that β-lactamase activity included in TEM-52 and the β-lactam antibiotic cefazolin could come into direct contact in the fish body leading to cefazolin degradation. These results thus clearly demonstrate that TEM-52 is active not only in vitro system but also in the fish body, i.e., in vivo following injections.
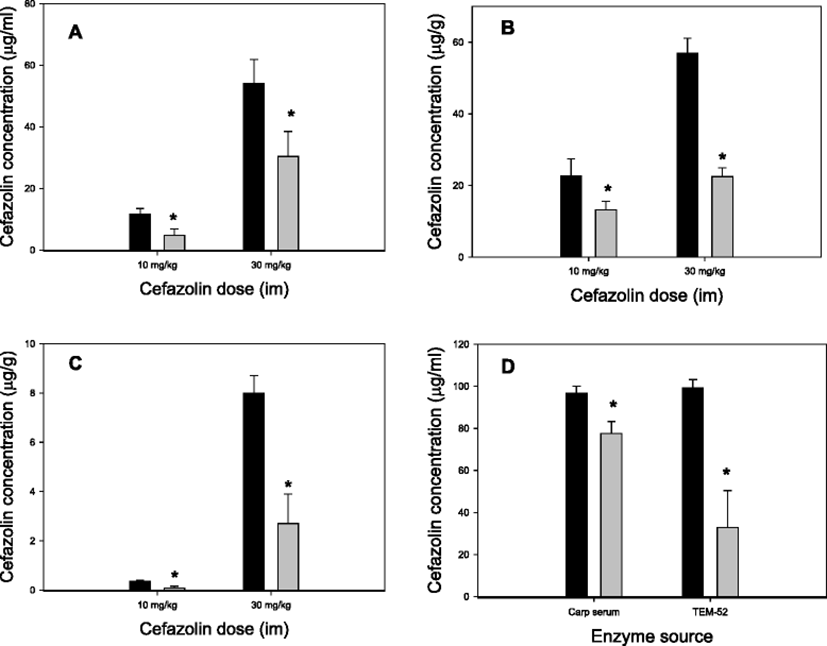
It was additionally checked whether TEM-52 was actually present in carp serum after intraperitoneal injection to the fish. For this, TEM-52 was injected and serum was sampled 1 h later for ex vivo degradation tests. After incubation of TEM-52-pretreated carp serum with cefazolin ex vivo, a significant reduction (p < 0.05) in cefazolin residue was observed (Fig. 1d, Carp serum columns). In a parallel test, it was observed that if a higher reduction of cefazolin concentration occurred, cefazolin was incubated directly with TEM-52 in the presence of naive serum (Fig. 1d, TEN-52 columns). Although it is difficult to compare directly because these two test conditions were not identical, a significant portion of intact TEM-52 enzymes seemed to reach in serum when blood was taken (1 h post-injection).
Because TEM-52 was active for cefazolin degradation in carp, it can be deduced that the administered enzyme must have been sufficiently absorbed from the peritoneal cavity. It is not well described in fishes whether there is an inhibiting barrier to proteins injected into the peritoneal cavity. However, the fact that peptides are systemically active following intraperitoneal injection in fishes (Hong and Secombe, 2009; Murashita et al., 2010) suggests a loose barrier within the peritoneum in fishes. In rats too, absorption of proteins from the peritoneum is comparable to that of isotonic solution (Flessner, 2005).
Conclusions
In summary, this study is the first attempt to seek a utilization of sorts of antibiotics degradation enzymes to depurate drug residues in animals. We observed that TEM-52, an enzyme active against a broad range of β-lactam antibiotics, can be a novel tool to remove residues from drug-treated fish bodies. It may be needed to expand tests against other β-lactam antibiotics and also in different fish species. There has not been any attempt in idea similar to this up to now. Various aspects of studies need to be carried out in order to establish practical usefulness. Safety of the test enzyme to humans is not known at all, for example.
Conclusively, however, a similar strategy will also be applicable to various classes of antimicrobials to which antibiotic-degrading enzymes have been reported.








