Background
Sardinella is a genus of fish in the family Clupeidae found in the Atlantic, Indian, and the Pacific Ocean. The paddle-shaped supramaxilla bones are major characteristics, which help distinguish Sardinella from other genera. Morphological characters distinguish Sardinella from all other clupeoid genera with the presence of two fleshy outgrowths on the hind margin of the gill opening (Whitehead 1985). According to FishBase (http://www.fishbase.org/), there are currently 22 recognized species in the genus Sardinella. Sardinella is important not only in marine food webs as a base consumer supporting tuna, seabirds, and marine mammals (Willette et al. 2011) but also in industry as the protein source with a low cost using as a bait for large fish or a feed in aquaculture.
Seven species in the genus Sardinella are currently known in Indonesian waters including Sardinella fimbriata, Sardinella gibbosa, Sardinella lemuru, Sardinella albella, Sardinella atricauda, Sardinella branchysoma, and Sardinella melanura, whose total catch in Indonesia reaches 278,600 tons in 2011 (MMAF 2012). Morphological identification in Sardinella is mainly characterized by their gill raker, pelvic scute, scales, and otolith (Homayuni et al. 2013; Bräger and Moritz 2016; Begg and Waldman 1999). However, species identification in the genus Sardinella is often hard for its broad geographical ranges, overlapping distributions (Willette et al. 2011) and morphological similarities (Sivakumaran et al. 1987) especially in larval stages (Ditty et al. 1994), which makes it difficult to manage the Sardinella resources in Indonesia.
In addition to the traditional morphological identification, the genetic information is now alternatively used for the species identification for its fast and exact results. The most widely used genetic markers are partial mitochondrial DNA sequences such as cytochrome C oxidase I (COI) or cytochrome B (CytB) (Palumbi et al. 1991; Ward et al. 2005; Vrijenhoek 1994). However, full mitochondrial genome sequences provide more information about its biogeographical or evolutional information than those fragmental sequences. Therefore, more than 5000 mitochondrial genomes have been deposited in GenBank database (www.ncbi.nlm.nih.gov) from 33,500 species identified based on morphological characteristics (www.fishbase.org).
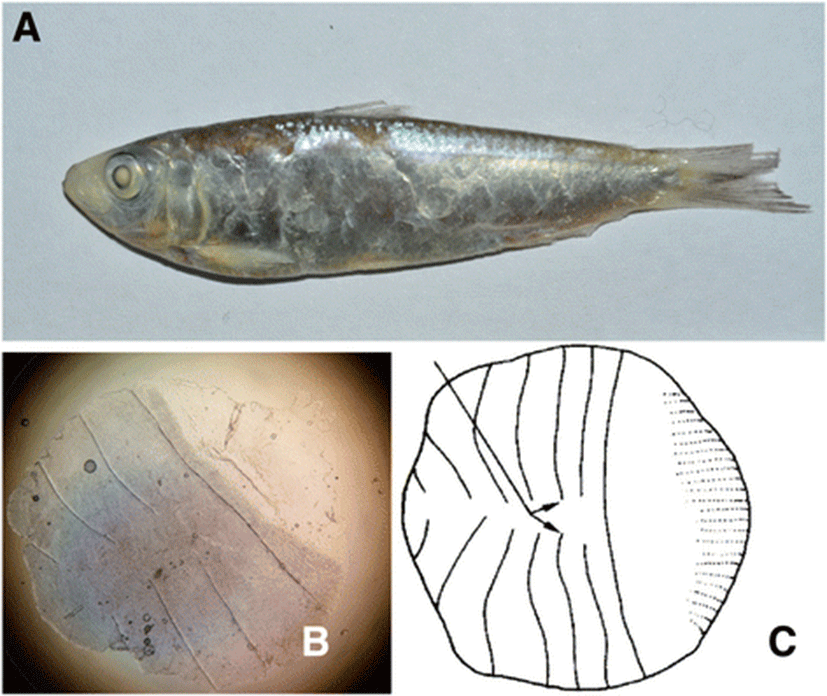
In this study, we report the Mauritian sardinella, Sardinella jussieu, for the first time in Indonesian coastal waters, which was collected from the Banten Bay. S. jussieu was previously reported only in the Western Indian Ocean, Taiwan, Hong Kong, and Vietnam (www.fishbase.org). Morphological characteristics of Sardinella jussieu are distinguished within other Sardinella species with the presence of black spot at dorsal fin origin and dusky pigmentation at caudal fin, total scute measurement which is 31, and vertical striae on a scale not meeting at center and no perforation on hind part (Whitehead 1985). After confirmation of the species by the molecular COI markers, its total mitochondrial genome sequence was determined by the combination of the traditional PCR methods and next-generation sequencing (NGS) techniques.
Methods
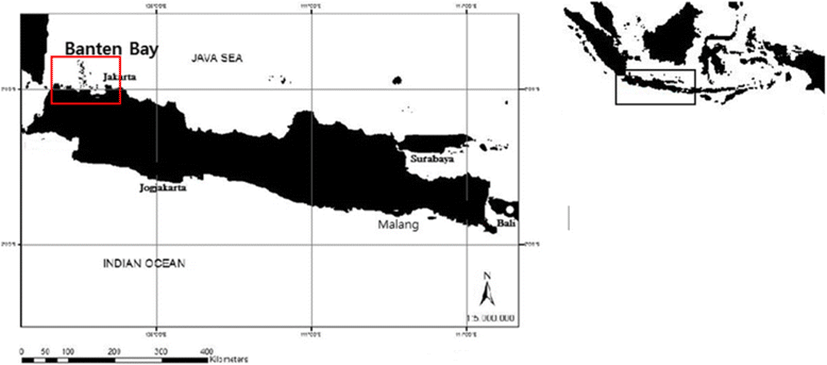
Five individuals of S. jussieu were collected in the Banten Bay, Indonesia (6° 0′ 50.00″–S 106° 10′ 21.00″ E), in January 2016 as the part of the regular fish survey (Fig. 1). Collected fish were directly stored in 96% ethanol and kept at − 20 °C until the further analysis (Knebelsberger and Stöger 2012). Morphological identification was made by their body shape, type of scale, fin feature, morphometric (i.e., standard length, body width, and head length), and meristic characteristic (total number of scutes) (Whitehead 1985; Strauss and Bond 1990).
Genomic DNA was extracted using an AccuPrep® Genomic DNA Extraction Kit (Bioneer) according to the manufacturer’s instruction. A small portion of tail fin was dissected, which was further homogenized by the TissueLyser II (Qiagen). Purified genomic DNA was quantified with nanoDrop (Thermofisher Scientific D1000), aliquoted, and stored at the − 70 °C for further analysis.
Two universal primer sets targeting cytochrome c oxidase I (COI) region, Fish F1 and Fish R1 (Ward et al. 2005), and targeting cytochrome b (cyt-B) region, GLUDG-L and CB2-H (Palumbi et al. 1991), were used to obtain the partial sequences of each gene, respectively (Table 1). The quality of all the primers used in this experiment was analyzed by the OligoAnalyzer 3.1 (http://sg.idtdna.com/calc/analyzer) and commercially synthesized by Bioneer Co. (Korea). Each PCR mixture (20 μL) contained 12.8 μL ultrapure water, 1 μL primer (0.5 μM, forward and reverse), 0.2 μL Ex Taq DNA polymerase (TaKaRa, Japan), 2 μL 10× Buffer, 2 μL dNTPs (1 μM, TaKaRa, Japan), and 100 ng genomic DNA as template. PCR was carried out under the following condition: initial denaturation step at 95 °C for 3 min, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 50 °C for 30 s, and extension at 72 °C for 45 s (COI target sequence) or 30 s (Cyt-B target sequence). The process was completed with a final extension at 72 °C for 10 min. Two PCR products targeting partial sequences of COI and Cyt B were then purified with AccuPrep Gel purification kit (Bioneer, Korea) and ligated into a cloning vector (Promega, USA), sequenced in both directions.
In order to obtain two large PCR products (~ 8 kb), two pairs of sequence-specific primer sets (CYT-F and CO1-R and CO1-F and CYT B-R) were designed based on the obtained partial sequences of each region (Table 1). Each PCR reaction (30 μL) contained 19.7 μL ultrapure water, 1 μL of each primer (0.5 μM), 0.3 μL Ex Taq Hot Start Version DNA polymerase (TAKARA, Japan), 3 μL 10× Buffer, 3 μL dNTPs (1 mM, Takara, Japan), and 100 ng genomic DNA as template. PCR was carried out with two-step PCR protocol for long PCR under the following condition: initial denaturation step at 94 °C for 3 min, followed by 30 cycles of denaturation at 98 °C for 10s, and annealing and extension at 68 °C for 10 min. The process was completed with a final extension at 72 °C for 10 min. Two large PCR products were pooled together in equal concentration and fragmented to ~ 350 bp in length by Covaris M220 (Covaris Inc.). TruSeq® sample preparation kit V2 (Illumina, USA) was used for the construction of a library from fragmented sequence and quality and quantity of the constructed library was measured using 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Sequencing was performed by Illumina Miseq platform (2 × 300 bp pair ends) (Illumina, USA).
Raw reads from MiSeq sequencer, with under Qv 20 and more than ambiguous nucleotides, were removed from raw read using CLC Genomic Workbench v 7.5 (CLC BIO Aarhus, Denmark). Mothür software was used to pairing forward and reverse sequence with more than 7 bp overlapped and without any mismatch. Paired sequence then assembled using Geneious R8 with minimum 20 bp of overlapping sequence and 100% overlap identity. Ambiguous sequences of the D-loop region were reconfirmed by the typical end-point PCR and with sequence-specific primers (Sard_F and Sard_R) and DNA sequencing of its PCR products by Sanger sequencing method (Table 1).
Results and discussion
As the result of morphometric measurements, we determined that the collected five fish were S. jussieu. Among the morphologically similar fish Sardinella species including S. albella, S. atricauda, S. fimbriata, S. marquesensis, S. sindensis, and S. gibbosa, the scale and pigmentation patterns are useful characteristics to identify species (Bräger and Moritz 2016; Strauss and Bond 1990). The average ratio of body depth (BD) to standard length (SL) of the collected samples was 27.5%, and total scute numbers were 31 (Table 2). Vertical striae on scales did not meet at center with no perforations on hind part of the scale, and the pigmented dorsal and caudal fins were also identified (Fig. 2). Those morphological characteristics suggested that the collected samples were S. jussieu. The most closely related Sardinella species, S. albella and S. gibbosa, are distinguished from S. jussieu in the presence of scale perforations (Table 3). Molecular identification of five Sardinella samples confirmed the morphological identification. The COI region of five individuals (652 bp) exhibited 100% sequence identity to Sardinella sp. (GenBank accession number: KJ566769) collected from the coastal water in Thailand and 99% to S. jussieu (GenBank accession no.: HQ231358) collected from the Philippines (Quilang et al. 2011). Based on the morphological characteristics and DNA sequence identity, we concluded that five Sardinella samples collected in the Banten Bay, Indonesia, were Mauritian sardinella, Sardinella jussieu.
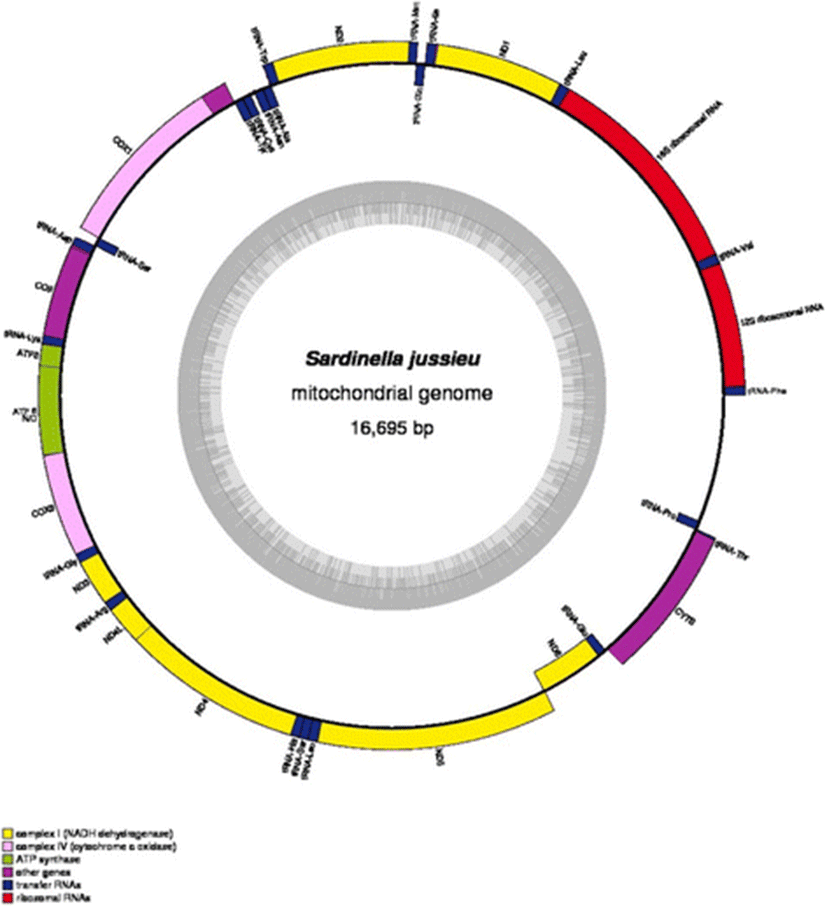
In order to have additional information of S. jussieu, the complete mitochondrial genome sequence was determined by the NGS and bioinformatic sequence assembly. Its mitochondrial genome was 16,695 bp in length consisting of 13 protein-coding genes, 22 tRNA genes, 2 ribosomal RNA genes, and the putative control region (Fig. 3). The base composition was 4415 A (26%), 4132 T (25%), 4900 C (29%), and 3248 G (19%). The purines and pyrimidines are A+T content (51%) slightly higher than G+C content (49%). The highest A+T content was observed in the putative control region (66%), which is similar to the other previous studies. The H strands encode 28 genes while the L strands encode 9 genes (Table 4). Among the protein-coding genes, three overlap nucleotides up to 10 bp, ATP8–ATP6, ND4L–ND4, and ND5–ND6, were detected. The transfer RNA gene pair tRNA−Ile–tRNA−Gln and t RNA−Thr–tRNA−Pro overlaps 1 bp as well. A total of 1292 bp of noncoding nucleotides are apparent in the S. jussieu with 1029 bp at putative control region, and 263 remains spread over 11 intergenic nucleotides; 68.3% (11.397 bp) of total mitochondrial genome sequence encoded 13 proteins and the size of each gene ranged from 168 bp (ATP8) to 1836 bp (ND5). Except for ND6, all protein-coding genes were encoded by H strand (Fig. 3). Although all 13 genes begin with typical start codon, ATG, there were several stop codons including typical ones such as TAA (CO1, COII, ATP8, ATP6, COIII, ND4L, ND5, CYTB), TAG (ND2, ND3, ND6, ND1), and exceptional AGA in ND4 gene (Table 4). Overlapping nucleotides were identified in three pairs of protein-coding genes (10 nucleotides for ATP8 and ATP6, seven for ND4L and ND4, and four for ND5 and ND6).
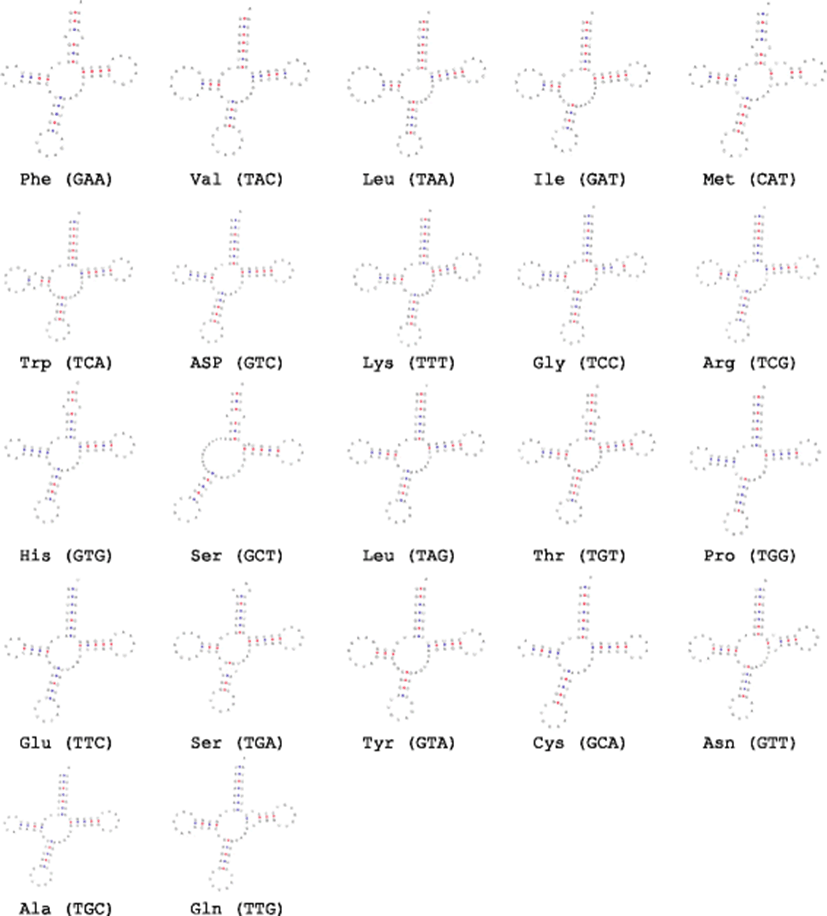
The mitochondrial genome of S. jussieu contained 22 tRNA genes (Fig. 4), which showed the difference in their sizes from 68 bp (tRNA–Phe) to 71 (tRNA–Gln). Fourteen tRNA genes encode in H strand and 8 genes encoded in L strand (Fig. 3). The 12S rRNA gene (951 bp) of S. jussieu was located between the tRNA–Phe and tRNA–Val, whereas 1686 bp of 16S rRNA was between tRNA–Val and tRNA–Leu. Twenty-one tRNA structures were predicted to have typical three arms except for tRNAser, which showed two arms. That result was also identified in the other Sardinella species (Lavoué et al. 2007). The putative control region of S. jussieu (1029 bp) was longest among three other Sardinella species including S. longiceps (958 bp) (GenBank accession number: NC033407), S. albella (986 bp) (GenBank accession number: NC016726), and S. maderensis (986 bp) (GenBank accession number: NC009587).
Total mitochondrial DNA sequence of S. jussieu showed 84–93% identity with those of currently known three other Sardinella species among which S. albella is the most closely related to S. jussieu (Fig. 5a). In order to know the better evolutional relationship of S. jussieu, its COI sequence was compared with those of the other 12 Sardinella species (Fig. 5b). As shown in the analysis by the full mitochondrial genomes, S. jussieu showed the most closely related to S. albella with 96% sequence identity. In fact, DNA sequence identity of two species S. albella and Sardinella gibbosa was too high to be distinct with each other in the COI region (Figs. 5b). Although morphological keys to discriminate two species were proposed, the post-pelvic ventral scutes and gill rakers number on a lower limb (Stern et al. 2016), S. albella and S. gibbosa frequently misidentified as shown in the COI barcodes. From the reason, it is required to compare full-length mitochondrial sequences of two species for the better classification. As the lowest sequence identity to other Sardinella species, control region of S. jussieu mitochondrial genome would be the good candidate to discriminate them.
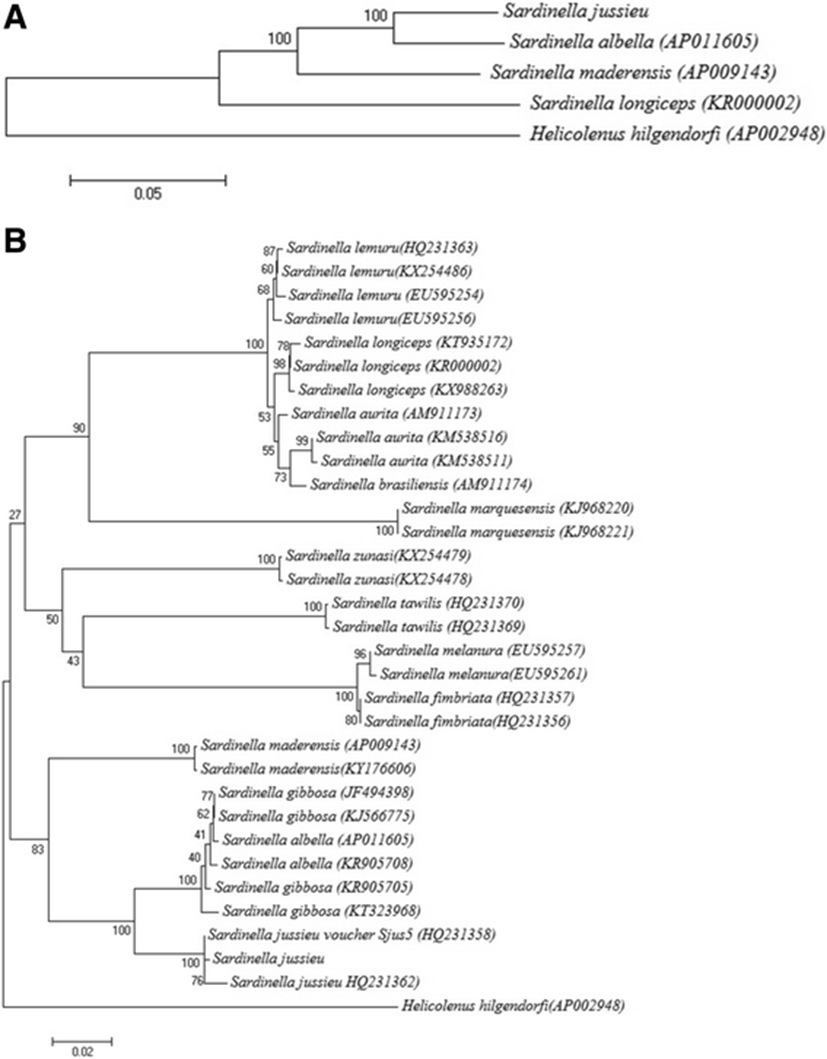
In this study, we identified that S. jussieu inhabits in Java island, Indonesia, as well as the two previously known Sardinella species, S. albella and S. gibbosa. Although S. jussieu is originally distributed in the western Indian Ocean from the western coast of southern India from Bombay South to Sri Lanka also Madagascar and Mauritius, the recent information is also caught in Taiwan (Hu et al. 2015), Hong Kong (Leung 1997), and the Philippines (Quilang et al. 2011). The result strongly supported that S. jussieu is more widely distributed than we have thought and the large-scale survey should be made to know the spatiotemporal distribution of four Sardinella species in Indonesia. We here reported the full-length mitochondrial genome sequence of S. jussieu collected from Java island, which would provide the important information for the scientific management of Sardinella species in Indonesia. We expect that more Sardinella species may exist in Java island and more information about the mitochondrial genome of the other unreported Sardinella species such as S. gibbosa would be a useful information for the molecular biological tools to discriminate different Sardinella species in Indonesia.
Conclusion
This study determined the complete mitochondrial DNA (mtDNA) sequence of S. jussieu in Java Island, Indonesia, for the first time. The mtDNA sequence is 16.695 bp in length and comprises the typical set of 2 rRNAs, 22 tRNA genes, 13 protein-coding genes, and putative control region. Mitochondrial genome structure of S. jussieu was identical to those in other Sardinella genus. Phylogenetic analysis using full mitochondrial genome exhibits that S. jussieu were most closely related to S. albella. However, comparison in the COI region showed that relationship between S. albella and S. gibbosa was ambiguous and determination of the complete mitochondrial DNA sequence of S. gibbosa is required for the better understanding of evolutional relationship between S. jussieu and those species. Those information would provide the basic information for the scientific management of Sardinella species in Indonesia.