Background
Metal-responsive transcription factor-1 (MTF-1; also termed metal-regulatory transcription factor-1 or metal-responsive-element-binding transcription factor-1) is a key transcriptional regulator playing pivotal roles in metal homeostasis and detoxification (Laity and Andrews 2007; Günther et al. 2012a). In addition to its fundamental role for homeostatic metal regulation, this multitasking transcription factor has also been known to be closely involved in cellular adaptation and protection against oxidative stresses through regulating the transcription of diverse genes related with host defense-related pathways (Günther et al. 2012a; Lichtlen and Schaffner 2001). They include metal reservation/detoxification (e.g., metallothionein, MT; the main target of MTF-1), metal ion transport (e.g., Zn or Cu transporters), iron homeostasis/anti-microbial responses (e.g., hepcidin), cellular redox homeostasis (e.g., selenoproteins and thioredoxin reductase), and glutathione biosynthesis (e.g., glutamate cysteine ligase) (Günther et al. 2012a; Lichtlen et al. 2001; Stoytcheva et al. 2010).
In a structural viewpoint, MTF-1 has been considered as a conserved transcription factor to possess six C2H2-type zinc fingers as the DNA-binding domain to recognize metal responsive elements (MREs) (Giedroc et al. 2001). As a cellular metal and stress sensor, the activity regulation of MTF-1 is generally characterized by the three successive steps, i.e., nuclear-cytoplasmic shuttling upon stress exposure, DNA-binding, and the interaction(s) with other coactivators to modulate the target gene transcription (Li et al. 2008). To execute the transcriptional regulation, MTF-1 binds to the specific site, called MRE (core sequence = TGCRCNC), in the promoter region of target gene (Günther et al. 2012a). Accordingly, the transcriptional expression of MTF-1 gene itself has been reported to be constitutive and not to be affected by heavy metal and other stressor treatments because its regulatory functions should be controlled mainly at post-translational levels (Auf der Maur et al. 2000; Bi et al. 2006). Structural scheme and functional context of MTF-1 above-described are believed to be widespread in the vertebrate lineage, although the majority of empirical information has come from mammalian MTF-1s.
However, in contrast to richness of knowledge on mammalian orthologs, molluscan MTFs have been narrowly explored barring only couples of previous reports (Qiu et al. 2013; Meng et al. 2015). Nevertheless, it is noteworthy that currently available molluscan MTF-1 sequences from public databases have suggested that mollusc species may show non-canonical features in their MTF-1 structures, which may differ from vertebrate orthologs. For instances, unlike vertebrate orthologs, molluscan MTF-1s often lack several typical motifs in presumed transactivation domains. Furthermore, a recent study has claimed that the nuclear-cytoplasmic shuttling, a key prerequisite step for vertebrate MTF-1s, might not always be an absolute precondition in certain mollusc species (Meng et al. 2015).
Pacific abalone, Haliotis discus hannai, is a highly valued seafood mollusc not only in Korea but also in other East-Asian countries. Intensive aquacultural operation for abalone farming using the marine net-cage system has been established in Korean aquaculture domain. During the last decade, the remarkable growth of abalone production in a quantitative term has been achieved (Park and Kim 2013). However, more recently, the sustainable progress of abalone culture has been considerably hurdled by the depressed productivity mainly in relation to frequent outbreaks of high mortality and physiological deformity in many abalone farms (Park and Kim 2013; Kang et al. 2015). Considering that abalone farming in Korea mainly relies on the net-cage facility installed in coastal areas, the heavy metals or other related pollutants contaminated in both water and sediments could be significant factors to provoke cellular toxicity and oxidative stress in farmed abalones (Kim et al. 2007).
However, despite its importance, adaptive or defensive functions to such environmental perturbations have been limitedly investigated in this abalone species, and almost no information has been available with respect to the coordinated regulations of genes involved in cellular pathways associated with metal regulation and oxidative stress responses (Kim et al. 2007; Lee and Nam 2016a). For this reason, understanding of MTF-1 from abalone species would be much useful to better comprehend orchestrated and coordinated regulations of host defense genes in this abalone species. Based on this need, this study, as a startpoint research, was aimed to characterize the genetic determinant of MTF-1, the superordinate regulator for diverse host defense genes, from the Pacific abalone (H. discus hannai). For this, we isolated and characterized the full-length complementary DNA (cDNA) encoding the abalone MTF-1, bioinformatically dissected its 5′-upstream regulatory region, and also scrutinized expression patterns of MTF-1 under both non-stimulated and stress-challenged (i.e., heavy metal exposure and heat shock) conditions.
Methods
Abalones (H. discus hannai) used in this study were experimental stocks maintained at Experimental Fish Culture Station, Pukyong National University (PKNU), Busan, South Korea. Abalones were maintained with semi-water recirculation system equipped with 3-ton capacity of rectangular culture tanks, in which the tanks were connected with protein skimmers, custom-designed mechanical filters, and 1-μm-mesh filter. Throughout the experiment, water temperature and dissolved oxygen were kept to be ranged within 20 ± 1 °C and 8 ± 1 ppm, respectively. Abalones were fed with frozen or dried seaweeds until 2 days before stress exposure treatments. Daily water exchange rate was about 20%, and in-tank wastes including feces and debris on the bottom were removed twice every day.
Based on the NGS-transcriptome analysis of the juvenile abalone tissues (unpublished data), partial NGS clones representing the significant homology to known animal MTF-1s were selected and assembled into a contig. In order to get full-length cDNA version, rapid amplifications of cDNA ends (RACE) at both 5′- and 3′-directions were carried out using total RNA isolated from a whole body sample and SMARTer® RACE 5′/3′ Kit (Clontech Laboratories Inc., Mountain View, CA, USA) according to the manufacturer’s instructions. Oligonucleotide primers used in this study are listed in Additional file 1: Table S1. The amplified fragments were sequenced and again subjected to contig assembly. Based on the assembled sequence in a contig, full-length abalone MTF-1 cDNA was re-isolated by RT-PCR amplification using the same total RNA aforementioned. Amplified RT-PCR products were directly sequenced at both forward and reverse directions to obtain a representative cDNA sequence for abalone MTF-1.
From the cDNA sequence, the 5′-upstream region of abalone MTF-1 gene was cloned by genome walking method. Using the genomic DNA prepared from an individual muscle, genome walking to 5′-upstream region was conducted with designated pairs of gene-specific primers and Universal Genome Walker® Kit (Clontech Laboratories Inc., USA) according to the manufacturer’s instructions. Amplified fragments were TA cloned into pGEM-T® easy vector (Promega, Madison, WI, USA), sequenced and assembled into a single contig. Afterward, the continuous, genomic fragment containing the 5′-flanking region was again PCR isolated from the genomic DNA template abovementioned and directly subjected to the sequencing to confirm the representative sequence of the abalone MTF-1 proximal promoter region.
With the ORF Finder program (https://www.ncbi.nlm.nih.gov/orffinder/), the open reading frame (ORF) of abalone MTF-1 was predicted and deduced amino acid sequence was obtained. Based on the homology search using NCBI BLASTx (http://blast.ncbi.nlm.nih.gov/Blast.cgi), sequence homology of abalone MTF-1 with its orthologs was examined. Parameter scores for the primary structure of MTF-1 were estimated using ExPASy ProtParam tool (http://web.expasy.org/protparam/). Multiple sequence alignment was done using CLUSTALW program (http://www.genome.jp/tools-bin/clustalw). Identification of putative zinc finger domains was carried out with Simple Modular Architecture Research Tool (SMART; http://smart.embl.de/). Predictions of potential nuclear localization signal (NLS) and nuclear export signal (NES) were conducted with cNLS Mapper (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi) and NetNES 1.1 Server (http://www.cbs.dtu.dk/services/NetNES/), respectively. The putative zinc finger-DNA binding regions from selected MTF-1 orthologs were subjected to molecular phylogenetic analysis using Molecular Evolutionary Genetics Analysis tool (ver. 7.0.21; http://www.megasoftware.net/). Putative transcription factor binding motifs in the abalone MTF-1 promoter were predicted with TRANSFAC® software (http://genexplain.com/transfac; GeneXplain GmbH, Wolfenbüttel, Germany).
Tissue distribution assay was conducted with two age classes of abalones. First, from 1-year-old immature juveniles (21.5 ± 4.1 g for total weight; n = 12), six kinds of tissues including the gill, gut, heart, hemolymph, hepatopancreas, and muscle (foot muscle) were surgically removed individually. For hemolymph, centrifugation (2500 rpm for 10 min at 4 °C) was carried out in order to collect hemocyte pellet. Second, from 3-year-old sexually mature adults showing a clear sign of ovarian and testicular maturation (96.4 ± 13.1 g for total weight; eight each for female and male), same tissue types abovementioned were obtained and additional ovary and testis were obtained. Upon surgically removed, biological samples were immediately frozen on dry ice and stored at −80 °C until used for RNA isolation.
In order to obtain developing embryos and early larvae, artificial insemination of sperm (from three males) to eggs (from eight females) was conducted by using the conventional induced spawning method including an air exposure and ultraviolet-irradiated seawater treatment. Insemination was made with wet-method at 20 °C and incubated at the same temperature until the end of sampling. An aliquot of developmental samples each consisting of approximately 20,000~30,000 embryos or larvae was sampled at six time points: just before insemination (unfertilized eggs), early cleavages (i.e., 2~8 cells stage; at 2 h post insemination; 2 hpi), morula (5 hpi), trochophore (12 hpi), early veliger (18 hpi), and late veliger (42 hpi), based on the microscopic examination. Upon sampling, embryos and larvae were also frozen on dry ice and stored at −80 °C until used. Two replicate samplings were carried out for each time point.
Two independent stress exposure experiments were carried out: one was with heavy metal exposure and the other with heat shock treatment. For heavy metal exposure, eight juvenile individuals (24.5 ± 3.6 g; approximately 1 year old) were assigned into one of five experimental tanks (70-L capacity containing 50 L of 1-μm filtered seawater at 20 °C) and acclimated to the tank conditions for 24 h before heavy metal treatment. The heavy metals used for exposure were of analytical grade reagents (Sigma-Aldrich, St. Louis, MO, USA). After 24 h, two tanks were treated with 0.02 mg/L (i.e., 20 ppb) and 0.1 mg/L (i.e., 100 ppb) cadmium (Cd), while two tanks with 20 and 100-ppb zinc (Zn) (Lee and Nam 2016a). Nominal concentration of the metal for each metal-exposed group was adjusted by using CdC12 or ZnCl2 stock solution. Remaining one tank was treated with only 1-mL distilled water that had been used for reconstitution of the metals (i.e., for non-exposed control). For each group, two replicate tanks were prepared identically. Treatment duration was 24 h. At the end of exposure, gill, hemocyte, hepatopancreas, and foot muscle were sampled individually from six randomly chosen individuals as described above.
On the other hand, for heat shock treatment, 22 individuals (21.1 ± 3.1 g; same-aged as above) were assigned into one of four 100-L tanks (two for heat-stressed groups and two for non-stressed groups) at 20 °C. Each tank was equipped with a custom-designed apparatus for mechanical filtration. After 24 h of acclimation period, water temperature of the two tanks (heat-stressed groups) was elevated using the adjustable thermostat-assisted aquarium heaters (400 W) with an increment rate of 1 °C/h. When the temperature reached 30 °C, the temperature was kept to be constant at 30 °C for additional 24 h. Samplings were made at 20 °C (just before thermal elevation), 25 °C (5 h after elevation started), 30 °C (10 h), 30 °C+12 h (12 h after reaching 30 °C; 22 h after elevation started), and 30 °C+24 h (24 h after reaching 30 °C; 34 h after elevation started). Four individuals were randomly selected from each tank to constitute eight individuals per temperature group at each sampling point. Tissues sampled were gill, hemocyte, hepatopancreas, and foot muscle. Meanwhile, non-stressed control groups were also identically prepared with heat-shock groups, but the temperature (20 °C) was kept to be constant until the end of experiment. At the same sampling point, the identical number of abalones (n = 4 per tank) was also obtained from non-stressed control groups. Temperature of each tank was confirmed to be ranged within ±0.5 °C. Dissolved oxygen levels were adjusted to be ranged from 7.5 to 8.5 ppm for all the experimental tanks. Abalones were not fed during stimulatory challenge experiments.
Total RNA was extracted using TriPure® Reagent (Roche Applied Science, Mannheim, Germany) and then purified using RNeasy Mini Plus Kit (Qiagen, Hilden, Germany) including DNase I treatment step. An aliquot (2 μg) of total RNA prepared was reverse transcribed into cDNA by using the Omniscript® Reverse Transcription Kit (Qiagen, Germany) including oligo-dT primer according to the manufacturer’s instruction. Synthesized cDNA was fourfold diluted with sterile distilled water, and an aliquot of 2 μL was included in a qPCR reaction as a template. The qPCR reaction was conducted with a LightCycler® 480 Real-Time PCR System and LightCycler® 480 SYBR Green I Master (Roche Applied Science, Germany), according to the manufacturer’s instructions. Thermal cycling condition for each gene (i.e., MTF-1 and normalization control genes) can be referred to Additional file 1: Table S1. Based on our preliminary study to evaluate candidate housekeeping genes for the normalization of RT-qPCR amplification (unpublished data; see also (Lee and Nam 2016a; Lee and Nam 2016b)), abalone ribosomal proteins L5 (RPL5; ABO26701) and L7 (RPL7; KP698945) genes were used as reference genes to normalize expression levels of MTF-1 transcripts in tissue samples (i.e., for basal tissue expression assays and stress exposure treatments), while RPL7 and RPL8 (KP698947) were used to normalize MTF-1 expression across developmental samples (embryos and larvae). Additionally, for heavy-metal exposure groups, the messenger RNA (mRNA) expression levels of MTF-1 were compared with those of MT (the known target gene of MTF-1) in order to examine whether or not there might be any positive or proportional relationship in the metal-mediated modulation patterns between MTF-1 and MT genes. PCR efficiency of primer pair for each gene was validated to be at least higher than 95% based on the standard curve prepared using a fivefold serial dilution of cDNA mix. For each cDNA sample, triplicate assays were carried out in an independent fashion.
Quantitative PCR-based MTF-1 mRNA expression levels across tissue types and developmental stages under non-stressed conditions were presented as ΔCt (Ct of the MTF-1 gene subtracted from the Ct of each internal control gene). On the other hand, differential expression levels of metal-exposed or heat shock-treated group relative to their corresponding non-stressed control groups were presented as the fold difference to the non-stressed controls by using the formula 2−ΔΔCt (Schmittgen and Livak 2008). Expression levels between or among groups were tested using Student’s t test or one-way ANOVA (followed by Duncan’s multiple ranged tests). Difference was considered to be significant when P < 0.05.
Results and discussion
The full-length cDNA of abalone MTF-1 was comprised of 48-bp 5′-untranslated region (UTR), a 1509-bp single open reading frame (ORF) encoding a polypeptide of 503 amino acids, and 582-bp 3′-UTR including a stop codon and 19-bp poly(A+) tail. A putative polyadenylation signal (AATAAA) was found at 21 bp prior to the poly(A+) tail (GenBank accession number; KT895224) (Additional file 1: Figure S1). The MTF-1 protein based on the deduced amino acid sequence was estimated to have 54.86 kDa of calculated molecular mass and 5.51 of theoretical pI value, respectively. Abalone MTF-1 represented quite a low sequence homology to its vertebrate and invertebrate orthologs where the maximum sequence identity at amino acid level was found to be only 27% with Biomphalaria glabrata (air-breathing freshwater snail; Gastropoda; Mollusca). Non-conservative feature without any appreciable sequence similarity to other MTF-1s is also found in the putative activation. Abalone MTF-1 was likely to possess only a shortened fragment (44-aa; pI = 4.06) presumed for the acidic domain and to lack almost entire region corresponding to the proline-rich domain and serine/threonine-rich domain of vertebrate MTF-1 orthologs (Additional file 1: Figure S2).
In contrast, the abalone MTF-1 was proven to share a high structural homology with other MTF-1s in the DNA binding domain (Fig. 1). All the MTF-1 proteins including the abalone MTF-1 (but except for the shortest Octopus bimaculoides MTF-1 having only five zinc fingers) were found to show highly conserved six C2H2-zinc fingers in their DNA binding domains. In their DNA binding domains, 12 cysteine residues and histidine residues were clearly conserved, in which the zinc finger domain of abalone MTF-1 showed the highest sequence identity (77%) with that of orthologue isoforms from Crassostrea gigas (Bivalve, Mollusca). However, the molecular phylogenetic analysis using the DNA binding domains has indicated that MTF-1s have been largely divergent in the molluscan phylum, which is apparently different form the monophyletic clustering of orthologs from Chordata phylum (Additional file 1: Figure S3). Functional partition of the zinc finger domains in abalone MTF-1 has been remained to be further characterized; however, from the mammalian studies, the region from the second to fourth zinc fingers have been proposed to constitute the core DNA binding domain while the first finger has been reported to serve as a metal-sensing domain (Bittel et al. 2000). Within each zinc finger, the His-X-Arg/Lys-X-His [H-X-(R/K)-X-H where X is any amino acid] motif has been known as a key site for zinc binding (Günther et al. 2012a), and it is preserved in the 1st to 5th zinc fingers for all the species examined in the present study. However, in the last finger (i.e., the 6th finger), the Arg/Lys residue inside the 5-amino-acid stretch was found to be conserved only in vertebrate MTF-1s. Meanwhile, all the molluscan MTF-1s showed a phenylalanine (Phe) as the first amino acid of the first zinc finger, whereas vertebrate and brachiopod orthologs possessed a tyrosine (Tyr) (Cheung et al. 2010).
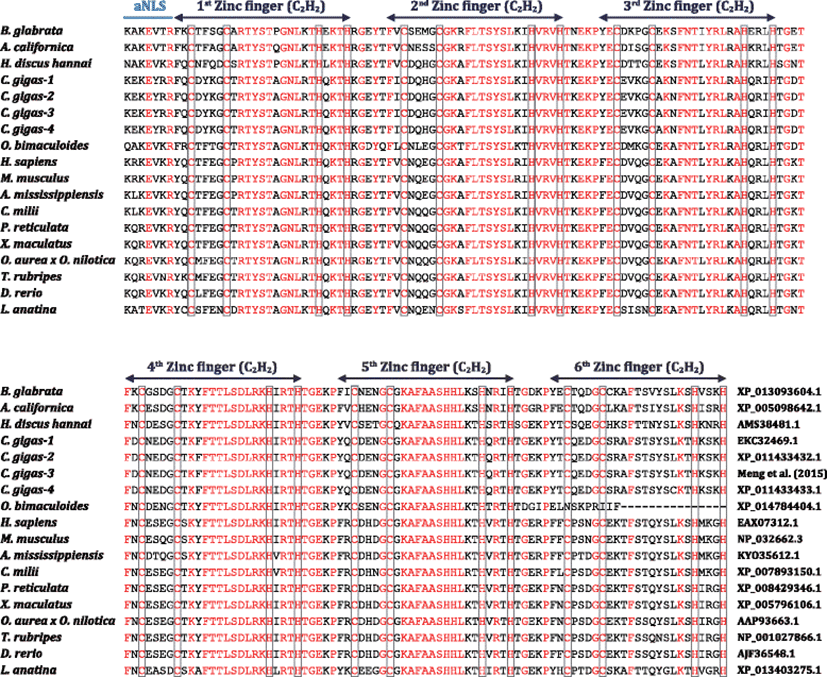
Abalone MTF-1 showed both conserved and unique features in the peptide linkers connecting zinc fingers. Conserved peptide linkers were found between 1st and 2nd fingers (Arg-Gly-Glu-Tyr-Thr), between 2nd and 3rd fingers (Thr-Lys-Glu-Lys-Pro), and between 4th and 5th fingers (Thr-Gly-Glu-Lys-Pro). On the other hand, unique linkers were found between 3rd and 4th fingers [Ser-Gly-Asn-Thr in abalone vs. Thr-Gly-Lys-Thr in vertebrates vs. Thr-Gly-(Glu/Asn/Asp)-Thr in other molluscs and a brachiopod species] and between 5th and 6th fingers [Ser-Gly-Glu-Lys-Pro in abalone vs. Thr-Gly-Glu-(Lys/Arg)-Pro in vertebrates vs. Thr-Gly-(Asp/Gly/Glu)-(Lys/Arg)-Pro in others]. Peptide linkers in multi-zinc finger domains have been reported to play roles in not only structural stabilization but also interfinger interactions for DNA-binding affinity of the zinc finger domains (Li et al. 2006). Particularly in the MTF-1, the linker between 1st and 2nd fingers has been known to be crucial in the zinc-sensing ability of the MTF-1 with regard to the formation of the ternary (MTF-1-zinc-DNA) complex for activating MT gene transcription (Li et al. 2006). Taking this into account, the abalone MTF-1 is thought to preserve a fundamental property of zinc-sensing function similarly with mammalian orthologs since it conserves a completely identical linker (i.e., the Arg-Gly-Glu-Tyr-Thr linker between 1st and 2nd fingers) with mammalian MTF-1s. However, the present abalone MTF-1 showed apparent dissimilarity with vertebrate orthologs in the linkers between 3rd and 4th fingers and between 5th and 6th fingers. Particularly because the linker between 3rd and 4th fingers has been reported to be important for the in vivo and in vitro sensitivity of zinc-dependent activation of MTF-1 (Li et al. 2006), this linker could be a good target for future studies to examine the potential difference in the linker-mediated zinc-finger function between abalone MTF-1 and mammalian/vertebrate orthologs.
In the abalone MTF-1, a putative auxiliary nuclear localization signal (aNLS) was identifiable in the front of the first zinc finger, as similarly with all other MTF-1 orthologs. Of seven amino acid residues to comprise the auxiliary NLS, the positions of two amino acids (3Glu and 7Arg) are conserved in all the MTF-1s examined (Fig. 1). However, due to the potential deletion in the putative acidic domain, no canonical nuclear export signal (NES) was observable in the abalone MTF-1 unlike vertebrate orthologs. Within a concept of MTF-1 activation (i.e., a nuclear-cytoplasmic shuttling function established in mammalian MTF-1), NLS and NES are responsible for balanced subcellular distributions of MTF-1 proteins (i.e., import and export, respectively) under both stressed and non-stressed conditions (Günther et al. 2012a). The NES motif in vertebrate MTF-1 is usually embedded in the acidic activation domain (Günther et al. 2012a; Cheung et al. 2010). However, the abalone MTF-1 does not show any typical NES motif, although a putative NLS motif is predicted in the front of the first zinc-finger domain that are also conserved all MTF-1s examined (i.e., conserved auxiliary NLS). The absence of acidic domain-embedded NES is not limited to abalone MTF-1, i.e., all molluscan MTF-1s are also likely to lack the NES at the corresponding region. Hence, our finding may suggest that molluscan MTF-1s could be different from mammalian MTF-1s in their subcellular localization control under both basal and stimulated conditions. A recent study with an oyster species (C. gigas) has claimed that the MTF-1 would primarily localize in the nucleus even under unstressed conditions and nuclear translocation might be uncritical for the activation of the oyster MTF-1 (Meng et al. 2015).
In addition, the typical motif of cysteine cluster (consensus sequence = Cys-Gln-Cys-Gln-Cys-Ala-Cys) that could be commonly found in the region immediately following the serine/threonine-rich domain of vertebrate MTF-1s was not detected in the abalone MTF-1 (Additional file 1: Figure S2). The cysteine cluster has been reported to be essentially necessary for metal-induced transcriptional activity and homodimerization of mammalian MTF-1 (Günther et al. 2012a; Günther et al. 2012b). Like with NES abovementioned, none of molluscan MTF-1 represents a canonical C-terminus cysteine cluster, suggesting that molluscan MTF-1s might have different mechanism(s) for metal-induced transcription (i.e., recruitments of transcriptional cofactor partners in the promoter/enhancer of target genes).
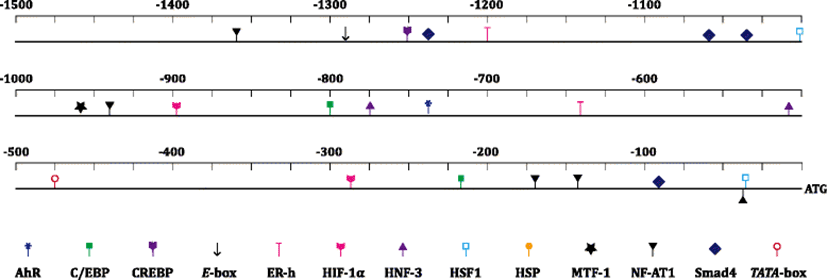
From the bioinformatic prediction, the 1691-bp 5′-upstream region from the ATG translation start site of the abalone MTF-1 gene represented various transcription factor binding sites (Fig. 2) (Additional file 1: Figure S4). The abalone MTF-1 promoter revealed a canonical TATA box (TATAAA) at −474 bp (from the ATG). Importantly, it represented a copy of MRE (TGCRCNC; −959 bp), suggesting the possible modulation of MTF-1 itself by heavy metal-driven cellular stressors, which is clearly inconsistent with the lack of MRE in many mammalian MTF-1 promoters (Auf der Maur et al. 2000; Bi et al. 2006). In addition, a xenobiotic response element (XRE; TNGCGTG; −736 bp) was predicted in the abalone MTF-1 promoter. XRE is an aryl hydrocarbon receptor (AhR)-targeted motif involved in the ligand (i.e., 2,3,7,8-tetrachlorodibenzodioxin (TCDD))-activated pathway to detoxify the effects of TCDD-related compounds. However, because invertebrate AhR homologues have been reported to lack the ability to bind TCDD directly (Hahn et al. 2006), the molecular mechanism on the potential interconnection between MRE/MTF-1 and XRE/AhR paths should be further explored. Nevertheless, recent mammalian studies have also highlighted multitasking roles of AhR in various signaling pathways associated with cell cycle control and antioxidant protection against oxidative stresses (Jackson et al. 2015). Besides, several transcription factor binding sites such as heat shock element (HSE; GAANRTTC; −1003 and −32 bp; targeted by heat shock factor (HSF)), hypoxia response element (HRE; RCGTG; −895 and −285 bp; by hypoxia-inducible factor-1 alpha (HIF-1α)), and other sites recognized by cyclic AMP response element binding protein (CREBP; TGACGY; −1253 bp) and nuclear factor for activated T-cells (NF-AT1; WGGAAA; −941, −169, and −33 bp) were predicted. All of these factors have been known to be related with stress responses of animals (Saydam et al. 2003; Dubé et al. 2011). Abalone MTF-1 promoter also revealed motifs that might be targeted by transcription factors generally known to be involved in development, signal transduction, cell proliferation and/or organ development. They included Smad4, hepatocyte nuclear factor (HNF), CCAAT-enhancer binding protein (C/EBP), E-box binding protein, and estrogen receptor (ER).
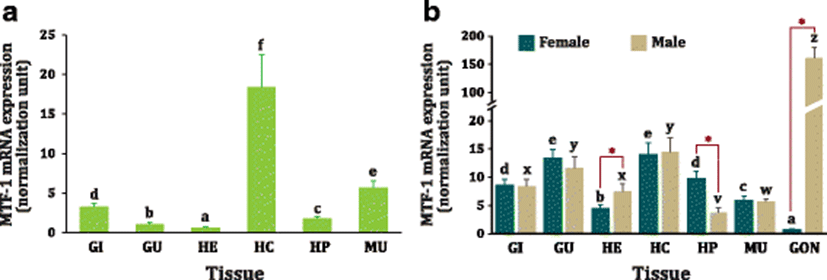
Based on the RT-qPCR analysis with immature juvenile abalones, MTF-1 mRNAs were detected in all the tissue types examined; however, basal expression levels were quite variable among tissues (Fig. 3). The ubiquitous detection of MTF-1 transcripts across all the tissues is not surprising when taken into account its housekeeping and fundamental roles in most cell types (Auf der Maur et al. 2000; O’Shields et al. 2014). The MTF-1 mRNAs were robustly expressed in hemocytes (P < 0.05), and this highest expression level was followed by muscles and gills, whereas the least mRNA expression was found in heart (P < 0.05). However, this expression pattern was not fully reproducible when measured with the sexually mature adults, although the broad pattern was consistent with findings from juveniles. Unlike in juveniles, the expression level of MTF-1 in the gut was found to be as high as that in the hemocytes in both female and male adults. More noticeably, matured adult abalones displayed a strikingly apparent difference in the MTF-1 expression in gonads where the extraordinarily high expression level was observed in testis while only minute expression in ovary (more than 150-fold difference; P < 0.05). Testis-predominant expression pattern of abalone MTF-1 in this study is similar with previous findings made in mouse (Auf der Maur et al. 2000) and hybrid tilapia (Oreochromis aurea × Oreochromis nilotica) (Cheung et al. 2010), collectively suggesting the possible involvement of MTF-1 in the male reproduction. Yet, the mechanism underlying the robust expression of MTF-1 in the abalone testes is currently unknown and open to hypothesize. In mouse, the reason for the high MTF-1 expression in testis has been explained by that sexually mature mice need to accumulate a large quantity of MTs in their testes (Auf der Maur et al. 2000; De et al. 1991). However, this hypothesis is unlikely to be adopted to this abalone species since the virtual increase of MT expression in sexually mature abalones has been observed in ovary rather than in testis (Lee and Nam 2016a), suggesting the molecular mechanism for the boosted expression of MTF-1 in abalone testis might be distinct from the ones in mammals. Hence, further study to monitor the MTF-1 expression in line with the testis development and maturation cycle would be valuable to get a deeper insight into this phenomenon. Besides the gonadal expression, matured abalones showed a sex-specific difference in the basal expression of MTF-1 in heart and hepatopancreas. Male abalones displayed higher expression in heart than females did whereas female abalones showed higher expression in hepatopancreas than males did. This finding is similar with a previous observation with zebrafish where males should have a higher MTF-1 mRNA expression in the heart than females (O’Shields et al. 2014). Although it has been still inconclusive for such a sex-related dimorphism, the response of zebrafish MTF-1 to Cd exposure has been reported to be gender dependent in some tissues (O’Shields et al. 2014).
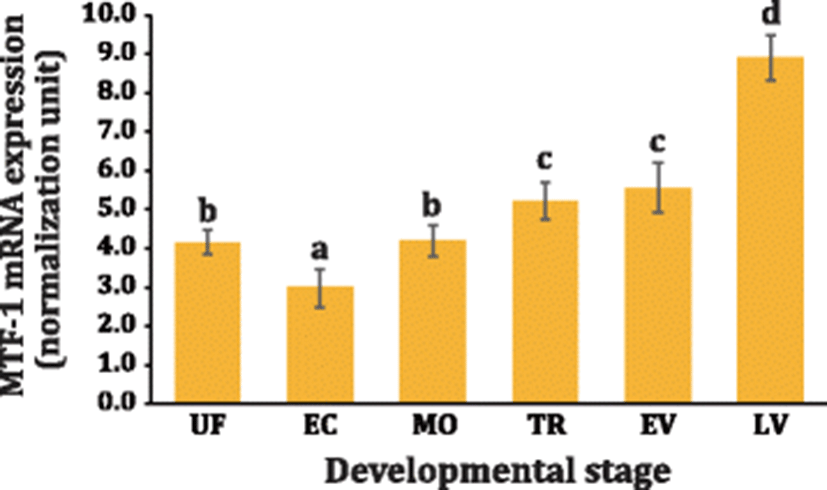
The MTF-1 mRNAs were found to be already present in unfertilized eggs based on RT-qPCR assay, which could be considered as a typical indicative sign of the maternal contribution of MTF-1 to offspring. Fine regulation of metal homeostasis should be one of the prerequisite requirements for developmental success of marine mollusc embryos that undergo external development in metal-residing sea water (Roesijadi et al. 1996; Jenny et al. 2006; Mao et al. 2012). The initial expression level was decreased down in early cleavage stages and rebounded to the initial level at morula stage. Although there was a trend toward increase of MTF-1 mRNAs with the developmental progress up to early veliger stage, the degree of upregulation was only modest. When the development progressed to late veliger stage, the mRNA expression level of MTF-1 was significantly elevated (P < 0.05) (Fig. 4). Developmental expression of MTF-1 in marine molluscan animals has not been yet characterized previously. However, the expression pattern observed in this study was generally in congruent with the anticipated roles of MTF-1 in embryonic and early ontogenic developments, as inferred from mammalian and teleostean cases (Günes et al. 1998; Chen et al. 2002; Chen et al. 2007). The expression pattern of MTF-1 during the development was also in agreement with the modulation pattern of its primary target (e.g., MT) in the same abalone species (Lee and Nam 2016a). Previously, the functional involvements of MTF-1 in the development and organogenesis have been highlighted by the lethality of “MTF-1-knockout” mice (Günes et al. 1998; Wang et al. 2004) and by induced inhibition of MTF-1 signaling followed by transcriptomic profiling in zebrafish embryos (O’Shields et al. 2014).
There was an apparent difference between the two metal ions in the modulation of MTF-1 gene expression. In overall, Cd induced potently the mRNA expression of MTF-1 while on the contrary, Zn repressed the MTF-1 expression (Fig. 5). Differential expression patterns of MTF-1 in response to Zn and Cd (20 and 100 ppb for both metals) were also dependent upon tissue types examined (gill, hepatopancreas, muscle, and hemocyte). In the gill, the exposure with 20-ppb Cd strongly induced the MTF-1 (P < 0.05) but higher exposure dose (100 ppb) did not give rise to the significant modulation of MTF-1 (P > 0.05). However, exposure with both doses of Zn significantly downregulated the MTF-1 in the gill (P < 0.05). On the other hand, in the hepatopancreas, both doses of Cd elevated the MTF-1 mRNA levels in a dose-dependent fashion (P < 0.05). Although the 20-ppb Zn exposure exhibited the small increase of MTF-1 mRNA levels in the hepatopancreas, 100-ppb Zn did not show any significant difference as compared to the level observed in non-exposed control (P > 0.05). Unlike in other three tissues, Cd exposure was unable to induce the MTF-1 expression in muscle tissue. In the muscle, Zn exposure depressed the MTF-1 mRNA expression (more significant downregulation of MTF-1 in the 100-ppb exposed group than in the 20-ppb exposed group). For hemocytes, the group exposed with 100-ppb Cd displayed a small, but statistically significant, increase of MTF-1 expression, while again, Zn-exposure resulted in the rapid downregulation of MTF-1 irrespective of exposure doses (P < 0.05).
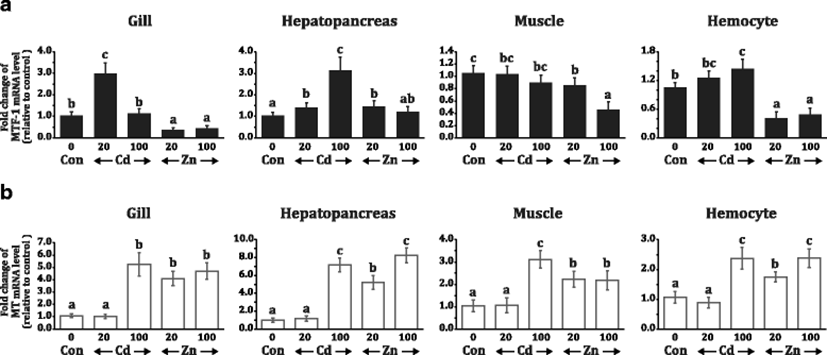
In contrast to the variable or opposite regulation of MTF-1 by Cd and Zn, the transcriptional response of MT (the main target of MTF-1) to the metal exposure treatments was relatively uniform. Further, unlike MTF-1 showing the downregulation upon Zn exposure in most instances, MT was found to be consistently upregulated by Zn in both 20 and 100-ppb exposure treatments (P < 0.05). Although the induced folds were variable among tissues, MT gene expression was unfailingly induced in all the four tissues by exposure treatments with 100-ppb Cd, 20-ppb Zn, and 100-ppb Zn, but not by 20-ppb Cd. Collectively, the modulation patterns upon metal exposure were apparently different between MTF-1 and MT genes, and the degree of Cd-mediated upregulation in each tissue was much higher for MT than MTF-1 (Fig. 5).
Previous studies have indicated that mammalian MTF-1 should be a constitutively expressed gene with a TATA-less promoter and that mammalian MTF-1s would not show any appreciable response to experimentally designed heavy metal and other stress factors. The plausible reason for the absence of metal (or stress) responsiveness has been explained by the lack of MRE motif in the mammalian (e.g., mouse and human) MTF-1 gene promoters (Auf der Maur et al. 2000; Bi et al. 2006). Hence, the MTF-1 activity in mammals is widely agreed to be largely induced at a post-translation level (Günther et al. 2012a; Smirnova et al. 2000; Saydam et al. 2001). However, in the present study, this abalone MTF-1 gene was proven to possess a putative MRE copy as well as a canonical TATA box in its promoter region. Accordingly, the present abalone MTF-1 gene displayed the Cd-mediated induction in multiple tissues (in gill, hepatopancreas and hemocytes, but not in muscles). Similarly, the Cd-induced expression of MTF-1 gene has also been reported in other cold-blooded animals such as zebrafish (Dubé et al. 2011; Cheuk et al. 2008) and carp (Ferencz and Hermesz 2009), which is obviously different from the principle for the MTF-1 regulation in mammals (Bi et al. 2006; Saydam et al. 2001).
However, the transcription of abalone MTF-1 was not induced by zinc except only a minute increase in the hepatopancreas; furthermore, Zn exposure even downregulated the mRNA expression of abalone MTF-1 in other tissues. This finding is similar with no induction of MTF-1 mRNA observed for zebrafish cells exposed to Zn (Cheuk et al. 2008), although Zn exposure in zebrafish was reported to be able to activate the nuclear translocation of MTF-1 from cytoplasm (Chen et al. 2007). On the other hand, a recent study with oyster C. gigas has shown that mRNA expression level of MTF-1 could be increased by Zn exposure, but the induced amount was only modest even treated with high dose of Zn (Meng et al. 2015). In the same report, authors have indicated that RNA interference of MTF-1 would result in the depression of Zn-mediated induction of MT (the known target of MTF-1), consequently confirming that MTF-1 is a superordinate regulator of the MT (Meng et al. 2015). However, in this study, the inducible pattern of MTF-1 was not in agreement with that of MT expression. Both Cd (inducer of abalone MTF-1) and Zn (non-inducer of abalone MTF-1) were found to be able to induce MT expression in all the tissues examined. Further, the quantitative relationship between MTF-1 and MT expression was not proportional. Currently, it has been widely agreed that Zn-mediated induction of MT could be achieved by direct binding of Zn to the MTF-1 fingers, and other metals such as Cd and Cu may not replace Zn in zinc finger binding (Schmittgen and Livak 2008; Chen et al. 1999; Zhang et al. 2001). Based on this, the Cd-mediated induction of MT gene might be considered as an indirect consequence. Possibly, Cd load might give rise to the release of Zn from MTs (also from other metalloproteins), and the increase of cellular Zn levels may activate MTF-1 through the Zn-binding. This zinc pool hypothesis can also be applied to the MT induction upon exposed to diverse oxidative stresses (Günther et al. 2012a). Hence, the controversy between MTF-1 and MT responses to zinc should be challenged in future study by examining cellular Zn concentrations (or tissue burden) under various Zn-exposure conditions.
Abalone MTF-1 was proven an early phase, heat-shock responsive protein as evidenced by the rapid modulation upon thermal increases. The MTF-1 mRNA levels in the gill were rapidly increased (fourfold relative to 20 °C; P < 0.05) as early as when water temperature reached 25 °C (Fig. 6). The expression level was further elevated when the temperature reached 30 °C (i.e., designated 30 °C+0 h) (P < 0.05). Afterward, the MTF-1 expression levels began to be decreased down with the continued incubation at 30 °C (i.e., 30 °C+12 h and 30 °C+24 h); however, the expression level at the end of thermal treatment was still significantly higher than that of 20 °C group (P < 0.05). On the other hand, in the hepatopancreas, the beginning of MTF-1 upregulation was more or less lagged as compared to that in the gill. Significant induction of MTF-1 in the hepatopancreas became evident from the 30 °C+12 h group, and the elevated expression level remained constant at 30 °C+24 h (P < 0.05). In muscles, the significant induction of MTF-1 was found at 30 °C+0 h and 30 °C+12 h but soon returned to the initial level observed at 20 °C group. Meanwhile, hemocyte displayed the rapid induction of MTF-1 mRNA at 25 °C; however, the expression dropped sharply at 30 °C+0 h in which the decreased expression level was even lower than that of 20 °C group (P < 0.05). Afterward, the MTF-1 expression rebounded at 30 °C+0 h group. Then, the rebound was followed by a further increase at 30 °C+12 h group (P < 0.05) (Fig. 6).
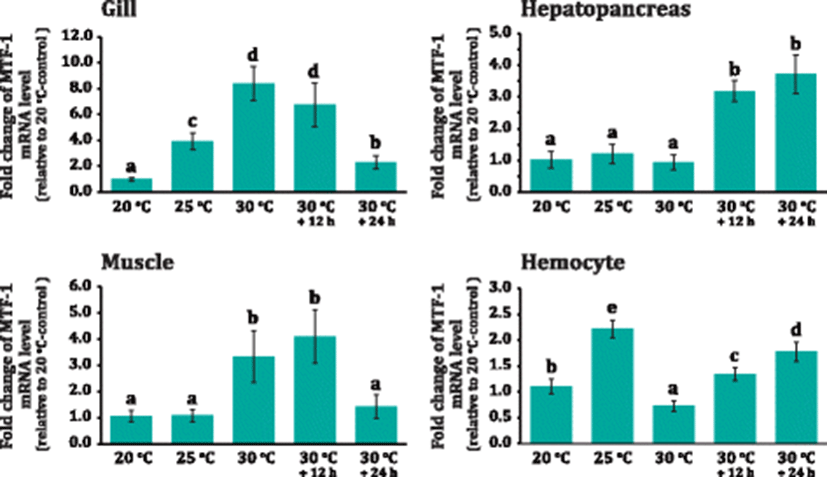
As aforementioned, the zinc pool sensing mechanism by MTF-1 upon exposure to stress factors might be adopted for the involvement of MTF-1 in host defense pathways against oxidative stress, since the abrupt and substantial changes of water temperature might be a causative factor to generate oxidative stress in poikilothermal invertebrates (Kim et al. 2007; Attig et al. 2014; Banni et al. 2014). Accordant with this explanation, the potential involvement of MT protein in the heat shock response has already been reported in this abalone species and also in other aquatic animals (Lee and Nam 2016a; Negri et al. 2013; Jarque et al. 2014). Within this context, the cold shock-induced MTF-1 has been reported in the common carp (Cyprinus carpio) brain, with an explanation that sudden temperature drop gave rise to the alteration of physiologically accessible Zn concentrations in that tissue (Ferencz and Hermesz 2008).
Meanwhile, a series of mammalian studies has proposed that MT and heat shock protein (HSP) genes might work in a non-cooperative way in their transcriptional responses to stress treatments. Experimental evidences for this proposal may include that (1) HSF should boost the activity of HSP gene promoter but hardly affect an MRE-containing promoter of MT gene upon heat shock and metal exposure, (2) heat shock-induced nuclear translocation of MTF-1 has been shown to be insufficient to activate a MT gene promoter, (3) diverse target gene searches for MTF-1 have been indicative of HSP genes as non-affected genes, and (4) MTF-1 has been likely to repress HSF-regulated genes through a direct protein-protein interaction (Lichtlen et al. 2001; Saydam et al. 2003; Uenishi et al. 2006). However, on the contrary, the present study has shown that the transcription of abalone MTF-1 could be directly activated by heat shock. Hence, our finding suggests that the thermal stress-mediated activation of MTF-1 in abalone may be achieved not only at a post-translation level (i.e., indirectly based on cellular zinc sensing abovementioned) but also at a transcriptional level (i.e., de novo synthesis). The presence of potential HRE motifs in the abalone MTF-1 gene regulatory region is also in congruent with the present hypothesis. Possibly, the activation of MTF-1 from the dual routes is likely to be advantageous for permitting the poikilothermal animals to prepare more efficiently antioxidant components (i.e., antioxidant enzyme genes containing MREs in their promoters as well as MT) against the oxidative stress caused by thermal fluctuations (Kim et al. 2007; Cho et al. 2009). However, further efforts are needed to get direct evidence on the cooperativity between HSF and MTF-1 in this abalone species.
Conclusions
Novel MTF-1 was isolated and characterized from a commercially important marine mollusc species, Pacific abalone (H. discus hannai). The abalone MTF-1 was found to share a conserved feature in zinc finger, DNA-binding domain with its orthologs; however, it represented non-conservative features in remaining other parts. Bioinformatic analysis of the 5′-upstream region has predicted diverse transcription factor binding motifs that are potentially related with metal regulation, stress responses, and development. Abalone MTF-1 was ubiquitously detected in various tissues, in which the highest expression level was observed in the testes of mature males. Abalone MTF-1 was expressed during the entire period of embryonic and early ontogenic development. From the heavy metal exposure, abalone MTF-1 was found to be Cd inducible (but not by Zn); however, the induced amounts were only modest as compared to that of MT. Abalone MTF-1 was highly modulated in responsive to heat shock potentially via both indirect zinc pool sensing and direct de novo transcription. Data from this study could be a useful basis to approach various researches regarding the stress responses in this abalone species particularly including detoxification of heavy metals and adaptation to thermal stresses.








