Background
Brown algal species of the genus Desmarestia (Desmarestiales) have a worldwide distribution (Guiry and Guiry 2017). Desmarestia species inhabit primarily the cold seawater of higher latitudes of both Northern and Southern hemispheres but are rarer in warm seawater (Graham et al. 2009). The genus includes free sulfuric acid-containing species, characterized by many branched or foliose macroscopic thalli with pseudo-parenchymatous cell structures (Yang et al. 2014).
Three species of Desmarestia have been reported from Korea, as D. ligulata H. Kawai, T. Hanyuda, D.G.Mülller, E.C.Yang, A.F.Peters and F.C.Küpper; D. tabacoides Okamura; and D. viridis (O.F.Müller) J.V.Lamouroux from Korea (Lee and Hwang 2010). Yang et al. (2014) revised the taxonomic relationship of Desmarestia species and suggested new combinations of the subspecies of D. dudresnayi and D. herbacea. On the species level, they established D. japonica from Japanese Desmarestia species.
Desmarestia japonica was recently established from Japanese Desmarestia species, based on molecular data and morphological characteristics (Yang et al. 2014). This ligulate species had been referred previously to D. ligulata in Japan, and its morphology was described by Okamura (1936) and Yoshida (1998) as D. ligulata.
Yang et al. (2014) stated that there was no evidence as to whether D. japonica occurred in Korea. Thus, there is a need to confirm the taxonomic entity and species boundaries.
In Korea, Desmarestia species show a restricted distribution in terms of ecological habitats. Thus, the National Institute of Biological Resources (Korea) established the scientific project on the distribution and genetic diversity of these rare species, and herbarium specimens of Desmarestia species have been deposited since 2007.
Herbarium specimens contain valuable information for genetic investigations (Nicholls 2009). DNA sequences from herbarium specimens can also provide the important molecular evidence to solve taxonomic controversies (Goff et al. 1994; Provan et al. 2008; Hughey and Gabrielson 2012; Saunders and McDevit 2012). However, most herbarium specimens have been found to not be in a suitable condition for molecular biological analyses. DNA degradation and contamination are still major limitations (Taylor and Swann 1994).
Many studies have attempted to overcome the limitations of herbarium specimens as molecular biological materials and to improve molecular tools for DNA extraction and amplification of target DNA regions (e.g., Taylor and Swann 1994; Meusnier et al. 2008; Prosser et al. 2016). Next-generation sequencing (NGS) was recently applied to extract genetic information from old herbarium specimens (e.g., Hughey et al. 2014; Suzuki et al. 2016).
For the selection of targeted DNA regions, shorter amplicons show higher efficiency in amplification. Thus, the universal DNA mini-barcode (cox1) with minimal length has been adopted for biodiversity analysis (Meusnier et al. 2008). However, this short length of the target DNA region could not solve the problem of contamination. During the preparation and conservation of specimens, many sources of contamination can be present. Epiphytic organisms on algal thalli may not be excluded completely during sample preparation. Many algal specimens have such epiphytic organisms, and thus, they may be included in any DNA analysis. Moreover, fungal and human DNA contamination can occur during conservation in herbaria.
In this study, we developed taxon-specific molecular markers for DNA barcoding of herbarium specimens of Desmarestia species deposited in the National Institute of Biological Resources (Korea). The taxon-specific primer pairs were designed for the amplification of DNA barcode regions (18S rDNA and cox1). We also report for the first time D. japonica from Korea.
Methods
We analyzed herbarium specimens deposited in the National Institute of Biological Resources, Korea (Fig. 1). The morphological characteristics of 21 specimens of Korean D. ligulata (Table 1) were measured. Photographs were taken with a digital camera (C-4040 zoom, Olympus, Tokyo, Japan) attached to a light microscope (BX50, Olympus). After the morphological reexamination, we cut a small piece (< 0.5 cm2) to minimize the damage from herbarium specimens to be used for DNA analyses. Images of specimens were obtained with a scanner (Epson, Seiko Epson Corp., Japan, Fig. 1).

Characters | Womersley (1987) (as D. ligulata) | Yoshida (1998) (as D. ligulata) | Yang et al. (2014) (as D. japonica) | Korean specimens (in this study) |
|---|---|---|---|---|
Color | Medium brown to greenish brown | – | Light olive brown | Light olive brown |
Height | Usually 20–50 cm tall | 60–100 cm tall | 0.6–1 (− 2) m tall | 40–67 cm |
Width of branches | With axis and primary laterals mostly 2–5 mm broad | 3–6 mm wide branches | 2–6 (− 20) mm wide branches | (0.7 −) 1.7–4 mm wide branches |
Holdfast | Discoid holdfast | – | Conical or flattened | Discoid holdfast |
Branching type | Fronds with frequent opposite branching to 3 orders | Opposite to ca. three times branching | Opposite, branches of limited growth branched to two to three times | Opposite, branched to two to three times |
Branch morphology | More or less linear but tapering apically and basally, dentate, and with a faint to conspicuous midrib (the axial filament) | Wide linear feather-like branches, tapering apically and basally with midrib | Branches stipitate and attenuate at base and apex | Branches stipitate and attenuate at base and apex |
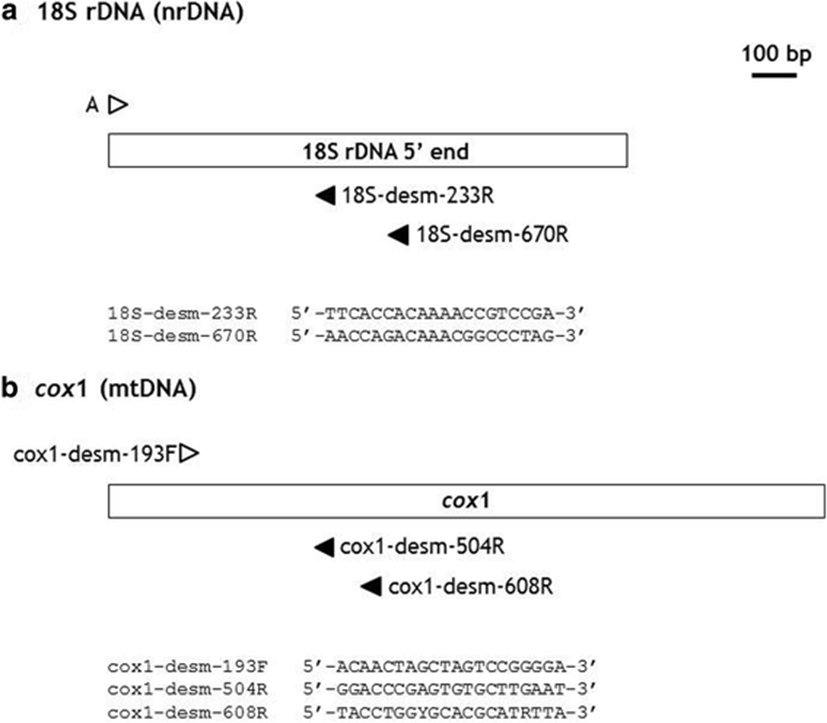
For the molecular analyses of the specimens, we used the reference sequences of Desmarestia species deposited in GenBank (NCBI, National Center for Biotechnology Information). To avoid contamination by fungi and other organisms, we developed taxon-specific primer pairs for the amplification of target DNA regions (Fig. 2, 18S rDNA and cox1). We selected a putatively conserved region among reference DNA sequences of Desmarestia species. Moreover, the DNA regions conserved with other organisms were excluded for the primer design as much as possible. The universal primer sets for 18S rDNA (A/SSUinR-1 in Lee et al. 2010) and cox1 (LCO1490/HC02198 in Folmer et al. 1994) were also tested for comparison.
The DNA extraction, polymerase chain reaction (PCR), and sequencing adopted the methods described in Lee et al. (2011). We isolated total DNAs from the subsampled herbarium specimens. We extended the incubation time of DNA extraction step (1 h). Moreover, the incubation times in washing step were also prolonged to improve the quality of DNA eluents. PCR conditions consisted of 3 min at 95 °C, 40 cycles of 30 s at 94 °C, 30 s at 50 °C, and 1 min at 72 °C, and a final 7 min extension step at 72 °C. Sequencing was conducted by a commercial service (Genotech, Daejeon, Korea), and the sequencing chromatograms were assembled with Sequencher 5.4.6 (Gene Codes Corp., Ann Arbor, MI, USA). The phylogenetic analyses were constructed using MEGA version 6 (Tamura et al. 2013). The neighbor-joining method and bootstrap analyses (2000 replicates) were used to reconstruct the phylogenetic tree. Molecular study about Desmarestia herbarium specimens had not been carried out in this lab previously. All reagents were in a sterile condition and stored in disposable plastic ware.
Results
Korean Desmarestia species showed a restricted distribution pattern, mainly on the northeastern coast mostly in subtidal habits (Lee and Hwang 2010). Because Desmarestia species live, herbarium specimens could be effective for the molecular investigation. We examined herbarium specimens deposited in NIBR collected from 10 years ago (Fig. 1). First, we selected samples previously identified as D. ligulata according to morphological resemblance (Yang et al. 2014). From the morphological examination, D. ligulata has proliferate pinnate-branched thalli and D. tabacoides typically has one or two wide-branched or unbranched foliose thalli (Table 1). In the case of D. viridis, this species was distinguished by much longer linear branched thalli.
We examined a total of 21 specimens identified as D. ligulata from the Korean coast. The thallus is light olive brown in color and, when exposed to air, becomes greenish brown. Korean specimens are up to 67 cm in height and have mostly three orders of branching. In the main axes and primary branches, branches were 2 mm wide, but in tall specimens, they were up to 4 mm wide. Gross morphology, with feather-like pinnate branching, was similar to that of Japanese ligulate Desmarestia species. Representative specimens showing morphological differences were also analyzed using molecular methods.
The universal primer set for 18S rDNA (Lee et al. 2010) produced fungal 18S rDNA sequences from the total genomic DNA extracts of herbarium specimens. The 18S rDNA sequenced showed high similarity with Agaricus bisporus var. bisporus (CP015465, 520/527(99%) from D. ligulata). However, we successfully isolated 18S rDNA (MF363011) and cox1 (MF363010) sequences of D. japonica from three specimens, using our taxon-specific primer pairs: NIBRAL0000000724 (Gangneung March 7, 2006), NIBRAL0000122790 (Gangneung May 8, 2009), and NIBRAL0000000705 (Goseong July 23, 2005).
Using forward primer A (Medlin et al. 1988; Lee et al. 2010), two reverse primers (Fig. 2a) produced PCR bands from DNA extracts of D. ligulata. The combination A/18S-desm-233R produced 213 bp, and A/18S-desm-670R amplified 650 bp of 18S rDNA without primer-binding sites. The three 18S rDNA sequences had the same sequence and 100% similarity with D. japonica (HE866912-HE866915, Yang et al. 2014). However, these 18S rDNA regions also had identical sequences with D. aculeata (HE866893-4), D. distans (HE866923), D. latifrons (HE866916), D. ligulata (HE866917-22), and D. muelleri (HE866924-5). Thus, these 18S rDNA sequences alone could not provide sufficient genetic information to discriminate interspecific relationships among Desmarestia species.
For the amplification of cox1 sequences (Fig. 2b, one forward and two reverse primers), the combinations of cox1-desm-193F/cox1-desm-504R and cox1-desm-193F/cox1-desm-608R successfully amplified the cox1 region of Desmarestia species. The primer pair of cox1-desm-193F/cox1-desm-504R showed high efficiency in amplification (272 bp excluding primer-binding sites). Thus, we used this combination to amplify cox1 from Desmarestia specimens.
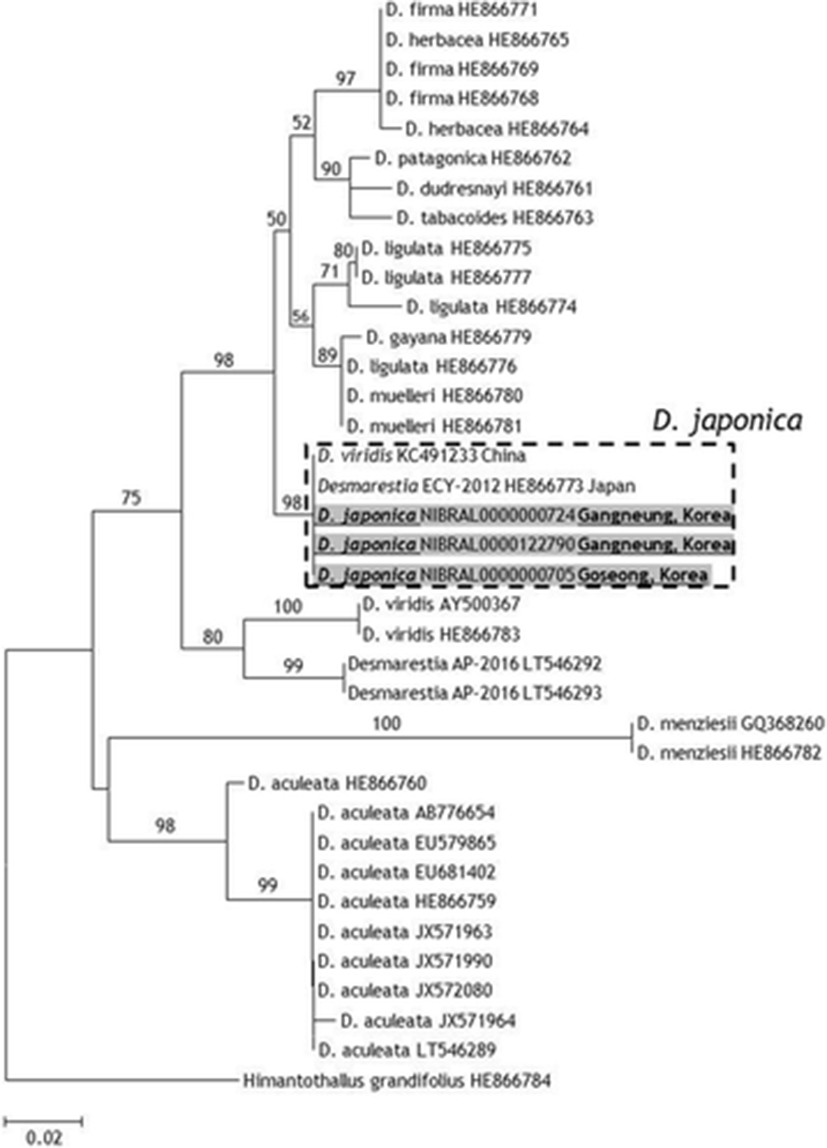
Korean D. japonica samples had the same cox1 sequence with Japanese D. japonica (HE866773 in Yang et al. 2014). A cox1 sequence reported from China as D. viridis (KC491233) also had 100% similarity with D. japonica. Because D. japonica showed below 97.4% similarity with other Desmarestia species deposited in the GenBank, this Chinese sample was likely misidentified (Fig. 3).
Discussion
The herbarium specimens examined from the Korean coasts have feather-like features and were smaller than the taller samples described by Yoshida (1998) as D. ligulata and Yang et al. (2014) as D. japonica (Table 1). However, they were similar in the color, branching pattern, and height of Japanese plants as well as Australian plants (Womersley 1987). Lamouroux’s (1813) illustration of D. ligulata showed that some primary laterals of the frond were dichotomous and some secondary laterals did not branch oppositely. However, we did not find such dichotomous branches within our Korean specimens whereas we did mostly observe opposite branching in secondary laterals.
Molecular phylogenetic studies of Desmarestia species were conducted to establish new species and to reconstruct the phylogenetic relationships (Tan and Druehl 1996; Yang et al. 2014). As a result, the key reference sequences of the 18S rDNA and cox1 region are available in GenBank. Thus, we selected these DNA sequences as the target regions for taxon-specific molecular markers of Desmarestia species.
DNA degradation in dried algal specimens and contamination are the major reasons for the failure of DNA analyses. The universal cox1 primer pair could not amplify the cox1 region from herbarium specimens of Desmarestia. In the case of 18S rDNA, fungal DNAs were amplified. Thus, a primer pair is required having high specificity and efficiency in amplifying the target DNA region from herbarium samples. In this study, we developed new primer pairs having short fragments of PCR to enhance the efficiency of amplification (Meusnier et al. 2008) and the specificity for the target plant samples (Fig. 2).
The primer pairs developed could successfully amplify 18S rDNA and cox1 regions from specimens of Desmarestia species. When the universal primers were used in the analyses, the samples showed no PCR band (cox1) or amplified fungal 18S rDNAs. The 18S rDNA and cox1 region could provide robust results for the finding of taxonomic entities of D. japonica. This report of D. japonica is the first regarding the distribution of D. japonica after the establishment of this species based on Japanese specimens (Yang et al. 2014).
The isolated 18S rDNA sequence of Desmarestia species could not provide a taxonomic resolution at the interspecific level and was not a suitable marker to analyze the taxonomic entities of Korean samples. The cox1 region has been selected frequently as a standard marker for algal DNA barcode use (Lane et al. 2007). In this study, the cox1 region provided suitable genetic information to examine the taxonomic entity of D. japonica from Korea. Yang et al. (2014) also found effective taxonomic resolution of the cox1 region, reflecting species delimitations among Desmarestia species and proposed the cox1 region as a potential barcode marker for the genus Desmarestia.
The overall morphologies of D. japonica specimens were variable in branching and branch width (Fig. 1, Table 1). Moreover, their morphologies were similar to those of D. ligulata.
In this study, we found D. japonica from herbarium specimens in NIBR using the taxon-specific primer pair (Fig. 2). These specimens were collected over 10 years ago and were first identified as D. ligulata based on morphological characteristics (Fig. 1). A Chinese cox1 sequence (KC491233) of D. viridis also showed 100% similarity in the cox1 region with Japanese D. japonica. These results indicate an extended distribution in Korea and China for D. japonica (Fig. 3). Consequently, a molecular taxonomic reexamination of the morphological resemblance among D. japonica, D. ligulata, and D. viridis is needed in future studies.
Conclusions
We developed taxon-specific primer sets to amplify the 18S rDNA and cox1 regions without contaminants (e.g., fungi and epiphytic organisms) and successfully isolated DNA regions from herbarium specimens over 10 years old. From these results, we confirmed the presence of D. japonica from Korea and China. We believe that the new molecular markers we have developed also provide useful information for DNA barcoding species of the economic seaweed Desmarestia.








