Introduction
Primary cell culture is a powerful tool commonly used to study the cellular properties and mechanisms of isolated cells in a controlled environment (Ager-Wick et al., 2018). Especially, primary culture of fish cells has been established in many fish species (Kim et al., 2018), it is also used as an ideal model for various study areas, such as virology, immunology, genetics, developmental biology, toxicology, medicine, biotechnology (Donato et al., 2008; Wang et al., 2010).
Cell lines offer the possibility of performing experiments in a controlled environment, independent of the complexity and variability of experiments in vivo (Jensen et al., 2013). For studies of host-pathogen interactions fish cell lines have become increasingly important, contributing to elucidation of constituents of innate immunity and of bacterial and viral virulence mechanism (Villena, 2003).
In order to conduct the fundamental research for the cell biotechnological applicability, we investigated the expression of immune-related gene in a new embryonic cell line (FGBC8) which was developed in our previous study (Kim et al., 2020). To achieve this aim, we performed quantitative real-time PCR (qRT-PCR) to observe the expression level of immune genes in FGBC8 cells following treatment with the several mitogens such as lipopolysaccharide (LPS), polyinosinic-polycytidylic acid (poly I:C), flagellin, and interferon (IFN)-γ.
Materials and Methods
A new olive flounder embryonic cell line called OFEC18-FGBC (FGBC8) was developed by our previous study (Kim et al., 2020). In order to subculture, cell culture media was arranged (L-15 medium containing 10% FBS and 2% AA) with 1% fish serum and 0.1% growth factor (basal fibroblast growth factor, Corning, Corning, NY, USA). Upon reaching 90% confluence, the cells were subcultured at a ratio of 2:3 as described our previous study (Kim et al., 2020).
FGBC8 cells (3.3 × 105 cells/well, 40 passages) were incubated overnight at 20°C in 6-well plates (Corning) and lipopolysaccharide (LPS; 10 and 50 μg/mL), Poly (I:C) (20 μg/mL), flagellin (5 μg/mL) or interferon (IFN)-γ (100 ng/mL) were added then incubated for 6 hours. Total RNA was isolated using TRIzol reagent and the RNA Extraction Kit (QIAGEN, Hilden, Germany). The first-strand cDNA synthesis was conducted using a first-strand cDNA synthesis kit (Roche, Basel, Switzerland) according to the manufacturer’s instructions.
qRT-PCR was performed following our previous study (Kim et al., 2020). Thermal cycling and fluorescence detection were performed using Fast SYBR Green Master Mix and SDS 7500 system (PE Applied Biosystems, Waltham, MA, USA). The immune gene expression analysis was conducted using the comparative Ct (2-ΔΔCT) method with 18s ribosomal RNA gene as a control. All samples analysis was performed in three replicates. The primers sequences used in this study are listed in Table 1.
Results were subjected to one-way analysis of variance (ANOVA) followed by Fisher’s protected least significant difference (PLSD) test using SPSS version 18 software. For all analyses, a p-value < 0.05 was taken to indicate statistical significance. All data represent the mean ± SD.
Results and Discussion
A new olive flounder cell line called OFEC18-FGBC (FGBC8) was developed by primary cell culture of blastula-stage embryos as described in our previous study (Kim et al., 2020). To explore the cell biotechnological applicability, the expression of immune related genes upon the mitogens such as LPS, poly I:C, flagellin, and IFN-γ were examined using quantitative real-time PCR.
Following immune stimulation, the expression of several immune-related genes was induced in FGBC8 cells. LPS is an essential component of endotoxin, also the release by heterophil granulocytes and monocytes/macrophages of many inflammatory cytokines, which have toxic effects on cells (Qin et al., 2009). Previous studies had been reported that the LPS induced the expression of fish IL-1β (Hong et al., 2003; Jiang et al., 2008). However, the PoIL-1β was strongly expressed by the flagellin stimulation, in this study (Fig. 1A).
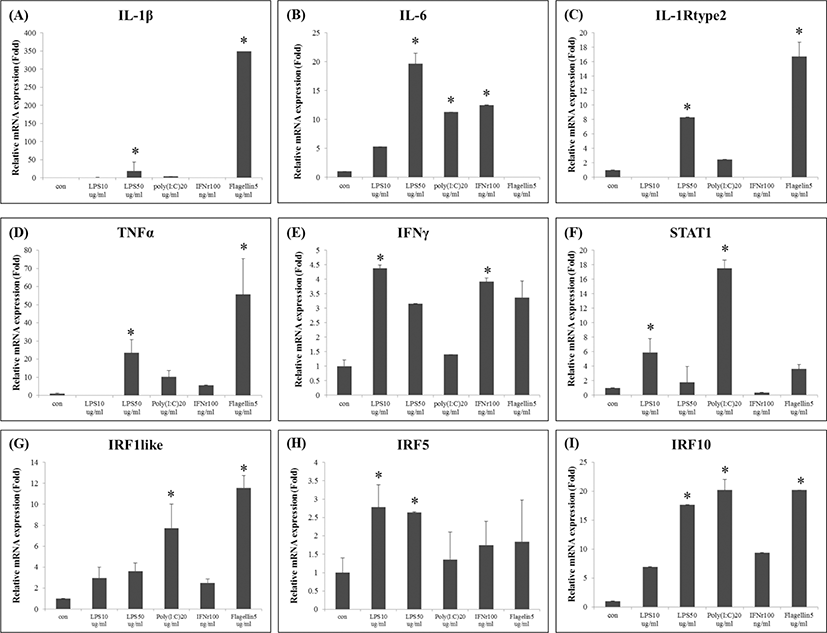
Flagellin is a component of the bacterial flagellum and induces pulmonary inflammation (Liaudet et al., 2003; Ritter et al., 2005). Recognition of flagellin by TLR5 triggers the MyD88-dependent signaling pathway, and the resulting activation of NF-κB induces the expression of proinflammatory genes, including TNF-α and IL-6 (Hwang et al., 2010). Interestingly, the lowest expression of PoIL-6 was observed after the flagellin stimulation (Fig. 1B), whereas PoIL-1Rtype2 and PoTNF-α were highly expressed by the flagellin (Fig. 1C, D).
During an innate immune response, IFN-γ is produced by natural killer cells following stimulation by IL-12 and/or IL-18 secreted by mononuclear phagocytes and antigen-presenting cells infected with intracellular pathogens (Jung et al., 2012). STAT1 is critical for the activity of IFNs and innate immunity (Park et al., 2008), moreover, the IRF-1 regulates IFN activity in fish (Collet & Secombes, 2002). Additionally, IRF10 is known to be involved in the function of leukocytes as well as the type 1 IFN response and the inflammatory response following pathogen infection in teleost fish (Suzuki et al., 2011). In this study, distinct expression level of immune genes such as PoIFN-γ, PoSTAT1, PoIRF1, PoIRF5, and PoIRF10 was observed in FGBC8 cells (Fig. 1E, F, G, H, and I) after all of immunostimulants.
In conclusion, our results demonstrate that FGBC8 cells will serve as a valuable research tool for further investigation of host-pathogen interactions and it is prospected that this cell line can be useful for cell biotechnological applications.








