Introduction
The innate immune system constitutes the first line of defense against pathogens penetrating marine organisms. Pattern recognition receptors (PRRs) initiate appropriate functions of the innate immune system and play vital roles in recognizing pathogen-associated molecular patterns (PAMPs) present on the surface of microbial pathogens (Kumagai et al., 2008). Among the PRRs, lectins are a group of carbohydrate-binding proteins that are primarily involved in pathogen recognition and triggering proinflammatory responses via protein-carbohydrate interactions (Vasta et al., 2011). Lectins are broadly classified into C-type, F-type, P-type, ficolins, pentraxins, and galectins.
Galectins are a conserved family of carbohydrate-binding proteins that include at least one carbohydrate recognition domain (CRD). These are widely distributed in mammals, birds, amphibians, fish, nematodes, sponges, and some fungi. In contrast to other lectins, galectins function in a Ca2+-independent manner and have two basic properties; the characteristic affinity for ß-galactosides, and the presence of a conserved CRD sequence motif. Evolutionarily conserved galectins have been classified into proto, chimera, and tandem-repeat types based on the structural differences of CRDs (Hirabayashi & Kasai, 1993). Proto-type galectins include one CRD with non-covalently linked homodimers. Chimera-type galectins comprise a C-terminal CRD and an N-terminal domain rich in proline and glycine. Tandem-repeat type galectins are embodied with two CRDs and a functional linker peptide that is required for joining the two domains (Vasta, 2012).
Galectin is a multifunctional protein involved in processes of cell development and migration, agglutination of erythrocytes and bacteria, and the regulation of adaptive immune responses. Previous studies reported that galectins act as mediators in developmental processes including cell differentiation, tissue organization, and regulation of immune homeostasis by binding endogenous (self) glycans (Leffler et al., 2002). Moreover, galectins can bind to exogenous (non-self) glycans present on the surface of viruses, bacteria, parasites, and fungi, as well as other PAMPs, as PRRs in the innate immune system (Chen et al., 2014; Shi et al., 2018; Wang et al., 2020).
Redlip mullets (Liza haematocheilia) belong to the Mugilidae family, which consists of more than 72 species from 17 fish genera distributed worldwide (Minos et al., 2009). Among them, redlip mullet is the only species being cultured and is regarded as a valuable aquaculture species in South Korea, China, Japan, and the Northwest Pacific Ocean. The statistics published by the Korean Ministry of Maritime Affairs and Fisheries have shown that collectively, mullets account for 8% of the total consumption and cultivation in Korea (Ministry of Maritime Affairs and Fisheries, 2020). Recently, the high mortality of the redlip mullet in Korea was attributed to Lactococcus garvieae, a gram-positive bacterium that causes green liver syndrome in these fish (Han et al., 2015). However, the knowledge of the host immune defense mechanism of redlip mullet is not known in adequate detail to control the outbreak of this disease. Therefore, identification and functional characterization of immune-related genes in redlip mullet are essential for their successful breeding.
In this study, we performed functional characterization and transcriptional profiling to investigate the potential effect of cDNA sequence of galectin-1 (LhGal-1) on the immune system of redlip mullet. Galectin-1 was identified from the redlip mullet (L. haematocheilia) cDNA database, and its spatial and temporal mRNA expression patterns were analyzed in tissues following immunogenic challenge with lipopolysaccharide (LPS), polyinosinic:polycytidylic acid (poly I:C) and L. garvieae. Additionally, the carbohydrate-binding ability of recombinant LhGal-1 was examined using lactose, galactose, and glucose. Furthermore, microbial binding assay and agglutination assay were performed for the functional characterization of LhGal-1 as a PRR.
Materials and Methods
All animal experiments were performed with the approval of the Institutional Animal Care and Use Committee at Jeju National University (approval no. 2018-0003). Healthy redlip mullets were obtained from the Sangdeok fishery in Hadong, South Korea, with average body weight and length of 146.5 ± 29.3 and 23.5 ± 1.7, respectively. Selected fish were reared at 20°C in 300 L tanks for seven days before the experiment.
Following anesthetization with tricaine mesylate (MS-222, 40 mg/L), five healthy redlip mullets were anatomized to collect the kidneys, spleen, gill, intestine, stomach, heart, blood, liver, muscle, skin, brain, and head kidney. A blood sample was drawn from their caudal vein using sterile syringes coated with heparin sodium salt (USB, Thermo Scientific, Waltham, MA, USA) and peripheral blood cells were collected by centrifugation at 3,000×g, 4°C for 10 min. All tissues were carefully isolated, frozen immediately in liquid nitrogen, and stored at –80°C until RNA extraction.
For the immune stimulation experiment, healthy mullets were divided into three immune stimulant groups and one control group; LPS (1.25 μg/g, from Escherichia coli 055:B5; Sigma-Aldrich, St. Louis, MO, USA), poly I:C (1.5 μg/g, Sigma-Aldrich), L. garvieae (strain: JJJN1, accession no.: CP026502.1, 1 × 103 CFU/μL) were prepared in 1 × phosphate-buffered saline (PBS). In different experimental groups, each fish was intraperitoneally injected with 100 μL of each stimulant, whereas an equivalent volume of PBS was injected to the fish of the control group. Subsequently, five fish were selected from each group and dissected 0, 6, 24, 48, 72 h post injection to isolate tissues, as described above.
Total RNA was extracted from the collected tissues (n = 5) to study the tissue-specific distribution of LhGal-1 in healthy fish, as well as upon immune challenge. RNAiso plus kit (TaKaRa, Kusatsu, Japan) was used to isolate RNA which was subsequently purified with RNeasy spin column (Qiagen, Germantown, MD, USA) according to the manufacturer’s instructions. The concentration and quality of purified RNA were determined with μDrop Plate (Thermo Scientific) at 260 nm and via 1.5% agarose gel electrophoresis, respectively. Thereafter, cDNA was synthesized using the PrimeScript™ II 1st strand cDNA synthesis kit (TaKaRa) in the final reaction volume of 20 μL containing 2.5 μg of RNA. The synthesized cDNA samples were diluted 40 folds and stored at –80°C.
For analyzing the tissue distribution and temporal distribution of LhGal-1 mRNA expression in different tissues following immune challenge, quantitative real-time PCR (qPCR) (TaKaRa Thermal Cycler Dice: TP850 Real-Time System, TaKaRa) was performed using cDNA as a template and gene-specific primers (Table 1). The gene encoding mullet elongation factor 1α (LhEF1α; accession no: MH017208) served as the internal control (Table 1). The 10 μL reaction mixture contained 3 μL of cDNA, 5 μL of 2× SYBR® Premix Ex Taq™, 0.4 μL of each primer (10 pmol/μL), and 1.2 μL of nuclease-free water. Each sample of cDNA was tested in triplicate. The following reaction conditions were used: initial denaturation at 95°C for 10 sec; 45 cycles of 95°C for 5 sec, 58°C for 10 sec, and 72°C for 20 sec; a final cycle of 95°C for 15 sec, 60°C for 30 sec, and 95°C for 15 sec.
Relative LhGal-1 mRNA expression levels were determined using the Livak (2−ΔΔCT) method (Livak & Schmittgen, 2001). The Ct values of all the samples were normalized to those of the LhEF1α gene. The Ct values in the tissues with the lowest expression of LhGal-1 were used for normalization when estimating the tissue-specific distribution. The data for the immune-challenged groups are shown as the relative fold-change compared to the PBS-injected group and calculated as the mean ± SD at a significance level of p < 0.05.
The redlip mullet cDNA database was constructed using the PacBio platform. All samples were sequenced using the Iso-Seq method (full-length mRNA sequencing technology) after determining the concentration and quality of RNA. For the library preparation, total RNA was isolated from blood, spleen, and head kidney and reverse transcribed into full-length first-strand cDNA using the Clontech SMARTer PCR cDNA Synthesis Kit (TaKaRa). The amplified cDNA was fractionated by size and converted into SMRTbell templates for sequencing. Finally, the full-length transcript isoforms were annotated using Blast2Go software (Liyanage et al., 2018). The cDNA sequence of LhGal-1 was identified using the Basic Local Alignment Search Tool (Altschul et al., 1990) available at the National Center for Biotechnology Information (NCBI) website and compared with other known orthologous genes sequences (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The open reading frames (ORFs) and translated amino acid sequences were determined using the online software ORF finder from NCBI (https://www.ncbi.nlm.nih.gov/orffinder/). Then, the NCBI Conserved Domain Search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), InterPro-EMBL-EBI (https://www.ebi.ac.uk/interpro/), and ExPASy-PROSITE (https://prosite.expasy.org/) were used to analyze the extrapolated protein domain sequences of LhGal-1. Besides, SignalP-4.1 server (https://services.healthtech.dtu.dk/service.php?SignalP-4.1) was used to ascertain the location and potential of the signaling peptide in the LhGal-1 sequence. Based on the sequence, the primers for cloning and qPCR were designed by Integrated DNA Technologies (https://sg.idtdna.com/pages), following MIQE guidelines (Bustin et al., 2009). Multiple sequence alignment was performed using CLC Main Workbench software version 8.0.1 (https://digitalinsights.qiagen.com) to determine the conserved domains and residues. Moreover, the Molecular Evolutionary Genetics Analysis (MEGA) software version 10.2 was used to perform phylogenetic analysis after using the neighbor-joining method with 5,000 bootstrap replicates to determine speciation and evolutionary relationships based on the similarities and differences within the molecular characteristics of different species (Kumar et al., 2018). Pairwise sequence alignment was performed using EMBOSS Needle pairwise sequence alignment software to determine the similarities and identities of the orthologs (https://www.ebi.ac.uk/Tools/psa/emboss_needle/). The 3D structural model of LhGal-1 was inferred from the amino acid sequence using Iterative Threading ASSEmbly Refinement (I-TASSER), which provides protein structure and function predictions based on the sequence-to-structure-to-function model (Roy et al., 2010).
The pMAL™ Protein Fusion & Purification System (New England Biolabs, Ipswich, MC, USA) was used to obtain a purified recombinant LhGal-1 protein (rLh-Gal-1). The ORF of LhGal-1 was cloned into the pMAL vector to append it to a sequence encoding the maltose-binding protein.
To clone the ORF of LhGal-1, target sequences were amplified via PCR with primers designed to include the corresponding restriction sites (Table 1). The 50 μL reaction mixture contained 5 μL of 10X ExTaq buffer, 4 μL of 2.5 mM dNTP, 10 pmol of each primer, 5 μL cDNA template, 0.2 μL ExTaq™ DNA polymerase (TaKaRa), and 33.8 μL nuclease-free water. The PCR was run on a thermal cycler (TaKaRa) under the following conditions: initial denaturation at 94°C for 5 min, 35 cycles of denaturation at 94°C for 30 sec, annealing at 59°C for 30 sec, and extension at 72°C for 40 sec, and final extension at 72°C for 7 min. Following PCR product purification, the pMAL-c5X vector (New England Biolabs) and inserts were digested using EcoRI and EcoRV followed by ligation using the DNA Ligation Kit (Mighty Mix) (TaKaRa). For the mass production of recombinant plasmid DNA, the E. coli DH5α competent cells were transformed and grown. The recombinant plasmid was extracted from the grown cells using the AccuPrep® Plasmid Mini Extraction Kit (Bioneer, Daejeon, Korea), and the samples were sent to Macrogen, Korea for sequencing.
The recombinant protein was overexpressed using isopropyl-β-thiogalactopyranoside (IPTG) induction. Briefly, transformed E. coli ER2523 cells were cultured in 500 mL Luria-Bertani (LB) broth containing 500 μL of ampicillin (100 μg/mL) and 0.2% glucose at 37°C. When the optical density at 600 nm (OD600) reached 0.6, IPTG (final concentration of 1 mM) was added to induce the expression of rLhGal-1 fusion proteins. The culture was further grown for 8 h at 25°C and 200 rpm. Next, the induced cells were harvested by centrifugation (3,500×g for 15 min at 4°C), washed twice with column buffer (20 mM Tris-HCL, pH 7.4 and 200 mM NaCl), and stored overnight at –20°C. For cell lysis, the cells were thawed in cold water and sonicated on ice. The lysate was centrifuged at 17,000×g for 30 min at 4°C. rLhGal-1 was purified using the pMAL™ Protein Fusion & Purification System (New England Biolabs) per the manufacturer’s instructions. Finally, the concentration of purified protein was measured using the Bradford assay and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed with 12% polyacrylamide gel to confirm the size and purity of the target fusion proteins (Bradford, 1976). The eluted protein was stored at –80°C until further analysis
The sugar-binding ability of rLhGal-1 was examined by performing an enzyme-linked immunosorbent assay (ELISA) and using three different carbohydrates as follows: α-lactose, D-galactose, and D-glucose (47287-U, G0750, G8270, Sigma-Aldrich). In brief, lactose, galactose, and glucose were each dissolved in carbonate-bicarbonate buffer (50 mmol/L, pH 9.6) to a final concentration of 100 mM. The 96-well microtiter plate was coated with the carbohydrate solutions and incubated at 4°C overnight. After washing three times with Tris-buffered saline with 0.1% Tween 20 (TBS-T), the wells were blocked with 5% skim milk in TBS-T. The plate was washed five times with TBS-T and treated with 50 μL of serially diluted rLhGal-1 or maltose-binding protein (MBP). After incubating for 1 h at 37°C and 10 rpm, the wells were washed three times with TBS-T, following which, 100 μL of mouse anti-MBP antibody (1:5,000, New England Biolabs) was added as the primary antibody. The plate was incubated at 37°C for 2 h and washed again with TBS-T. It was then incubated with the secondary antibody (100 μL goat anti-mouse IgG horseradish peroxidase conjugate; 1:3,000; YOUNG IN Frontier, Seoul, Korea), under the same conditions used for the primary antibody. Finally, the plate was washed five times with TBS-T and incubated with tetramethylbenzidine solution at room temperature for 5 to 10 min in the dark. For terminating the reaction, 50 μL of stop solution (1 M H2SO4) was added to each well and the absorbance was measured at 450 nm using a Multiskan Sky Microplate Spectrophotometer (Thermo Fisher Scientific).
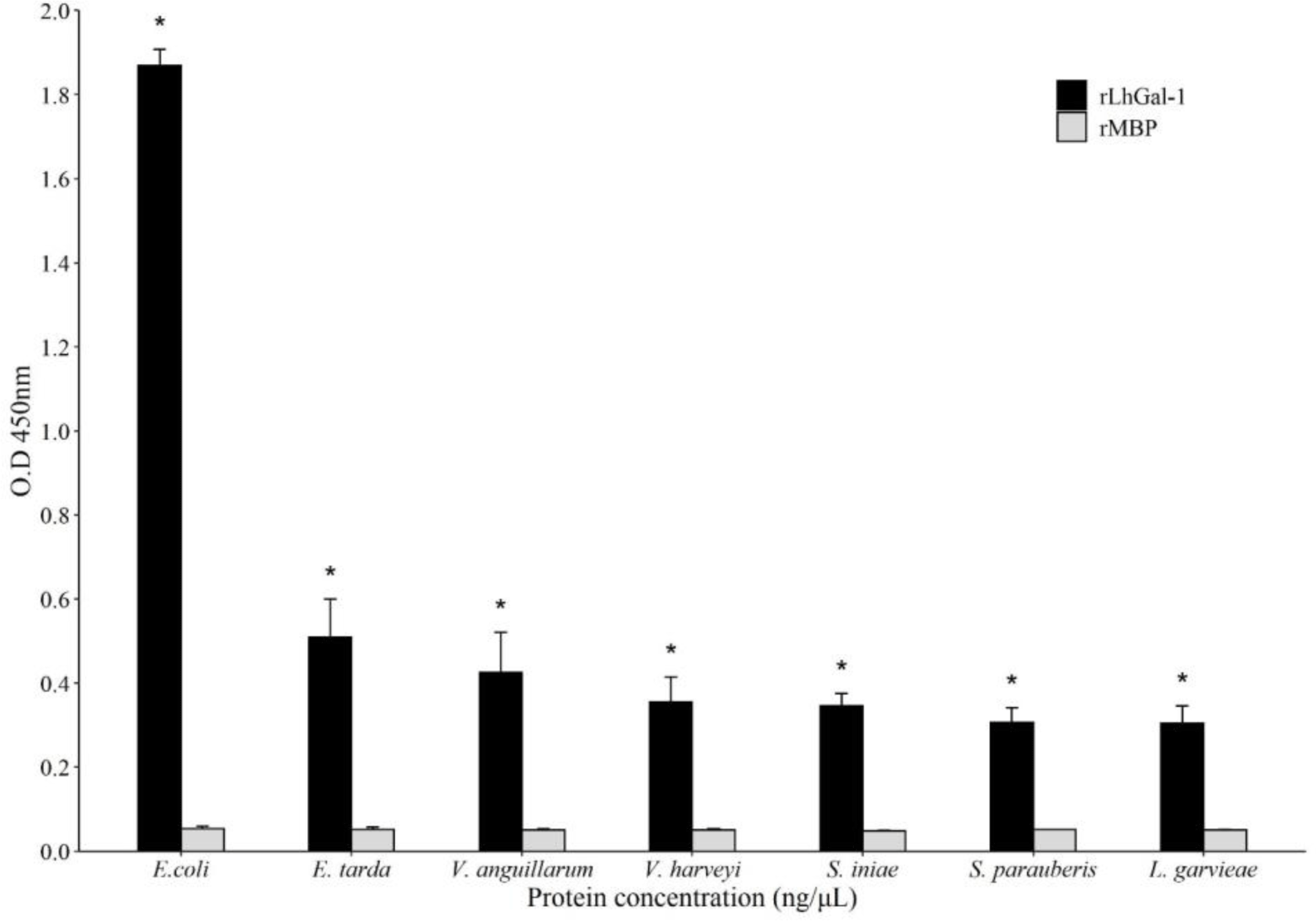
To determine the microbial binding affinity of rLhGal-1, ELISA was performed with seven bacterial strains as follows: three gram-positive (G+) species (L. garvieae, Streptococcus iniae, and Streptococcus parauberis) and four gram-negative (G–) species (E. coli, Edwardsiella tarda, Vibrio anguillarum, and Vibrio harveyi). Briefly, the seven bacteria were cultured in appropriate media overnight and harvested by centrifugation; LB broth was used for growing E. coli and V. anguillarum, and brain heart infusion broth with 1.5% NaCl was used for growing L. garvieae, S. iniae, S. parauberis, E. tarda, and V. harveyi. Harvested cells were washed twice and resuspended in PBS until an OD600 of 1 was achieved and diluted with carbonate-bicarbonate buffer (50 mmol/L, pH 9.6) to achieve the bacterial count of 1 × 108 cells/mL. The 96-well plate was coated with 100 μL of bacterial cell suspension diluted in coating buffer (1 × 107 cells/well) and incubated overnight at 4°C. ELISA was performed as described for the sugar-binding assay.
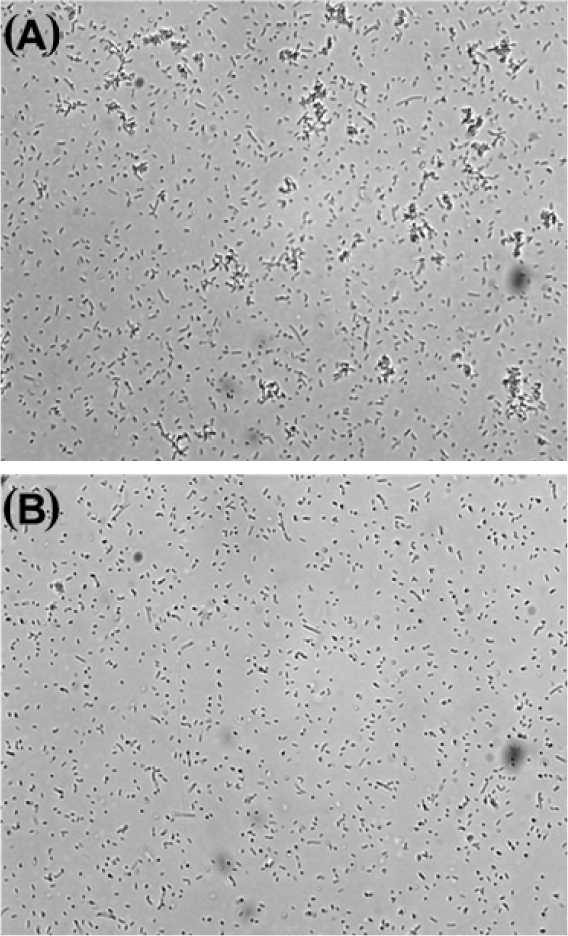
To examine the agglutination ability of rLhGal-1, the agglutination assay was performed using E. coli. Briefly, E. coli was cultured in LB broth at 37°C overnight and harvested by centrifugation. Harvested cells were washed twice with TBS (20 mM Tris, 137 mM NaCl, pH 7.8) and 50 μL of cell suspension was plated into the individual wells of a 96-well plate (5 × 106 CFU/mL). Each well was treated with 10 μg of rLhGal-1 and incubated at 25°C for 1 h. The control group was treated with an equal amount of MBP. Bacterial cells were observed under a light microscope to determine agglutination. All assays were performed in triplicate.
All experiments were performed in triplicates and the data are presented as the mean ± SD. The Student’s t-test was used to compare the mean values of control and challenge groups and determine the statistical significance of the results. One-way analysis of variance was used to examine the significant differences within the groups. Results with p < 0.05 were considered statistically significant.
Results and Discussion
LhGal-1 was identified from the constructed redlip mullet transcriptome database and characterized by sequencing and using several bioinformatics tools. The length of LhGal-1 (GenBank accession No: MW147017) ORF was 408 bp and encoded a protein of 135 amino acids with a predicted molecular weight of 15.31 kDa, and a theoretical isoelectric points of 4.89 (Fig. 1). The signal 4.1 server predicted a lack of signal peptides in the whole amino acid sequence, suggesting that these molecules might be secreted via a non-classical secretory pathway similar to other galectins (Boulianne et al., 2000).
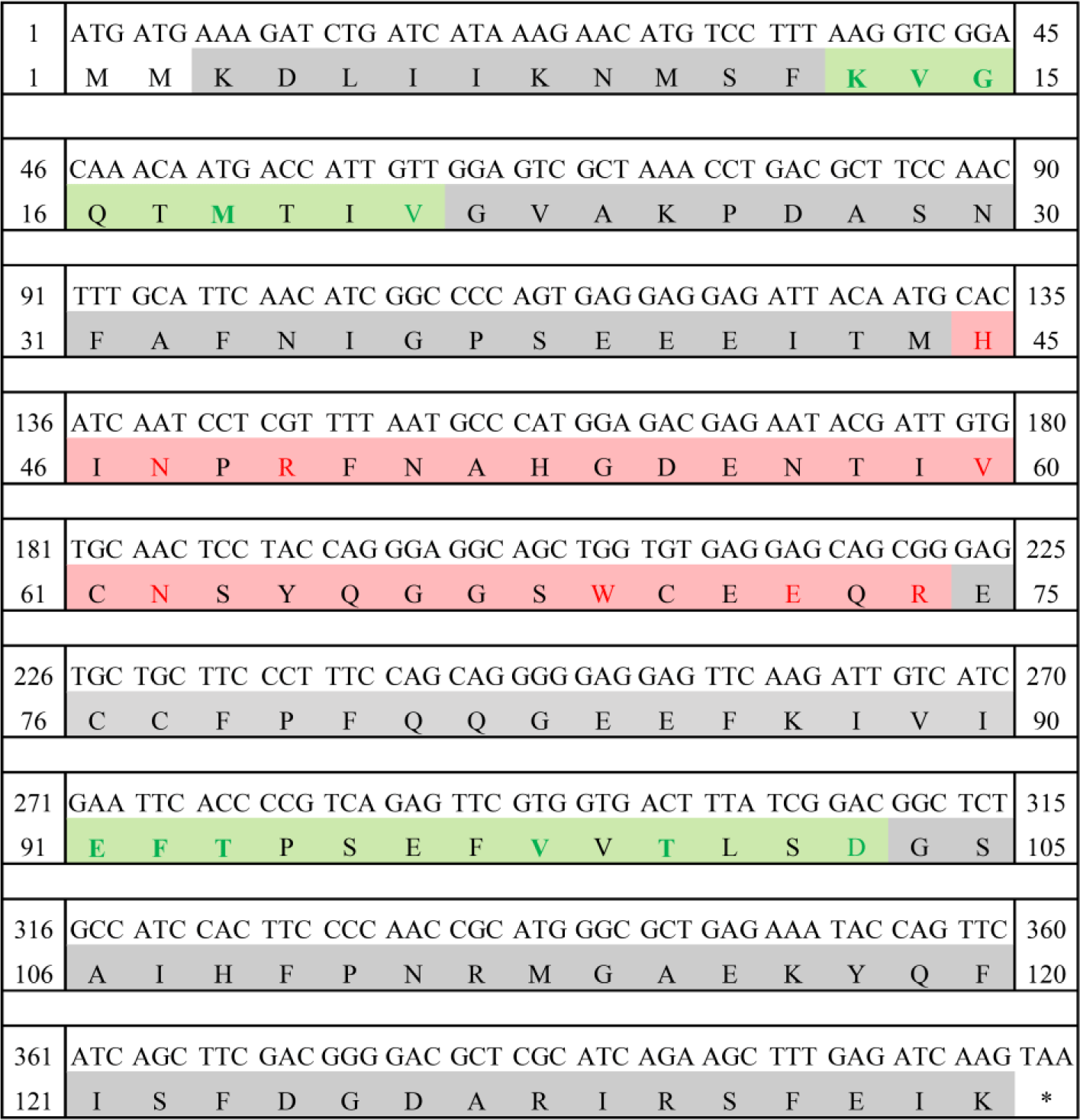
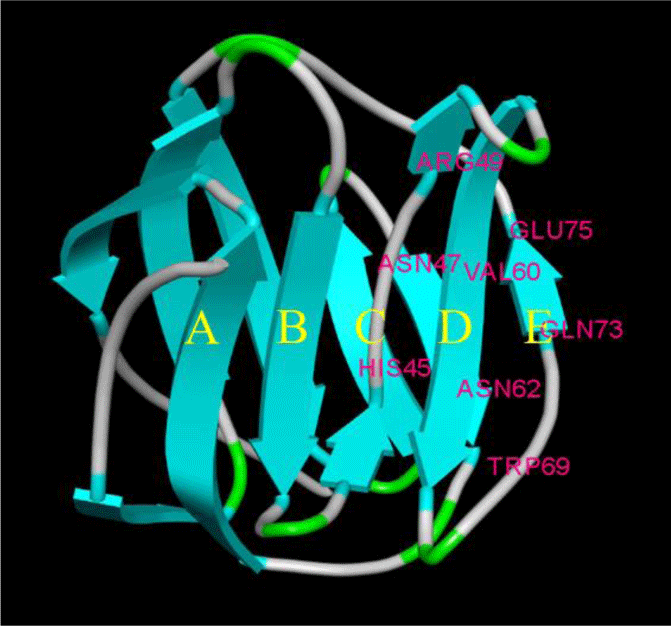
The amino acid sequence of LhGal-1 harbors a single GLECT CRD (Letunic & Bork, 2018). LhGal-1 sequence can be considered as the proto-type of galectins (Vasta, 2012). According to our InterPro analysis, GLECT domains of LhGal-1 contained eight conserved residues of sugar-binding pockets for β-galactoside (45H, 47N, 49R, 60V, 62N, 69W, 72E, and 74R) (Fig. 2). Although LhGal-1 has four cysteine residues (61C, 70C, 76C, and 77C), a disulfide bond was not identified by the ScanProsite software (Fig. 2). The predicted 3D model of LhGal-1 revealed that it is folded as a β sandwich comprising two anti-parallel β-sheet bundles (Fig. 2). Furthermore, five strands of each β sheet bundle were slightly bent and shaped to generate a concave and a convex side. All the main residues for carbohydrate binding were positioned on the concave side of the structure. This groove-like formation may allow the holding of linear tetra-saccharide long enough to form a sugar-binding pocket (Leffler et al., 2002).
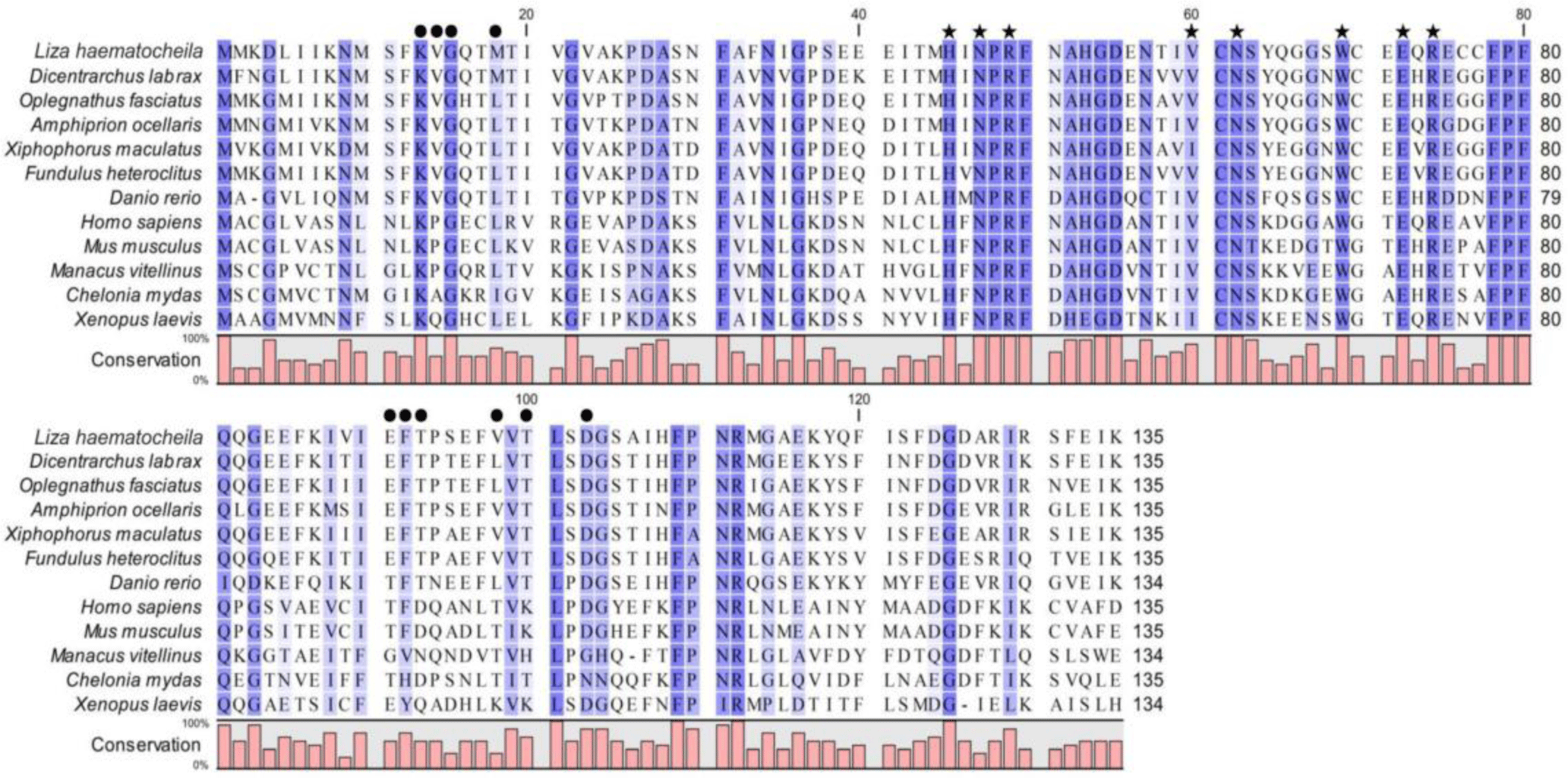
Multiple sequence alignment showed that LhGal-1 harbored highly conserved motifs within the CRD (H-NPR, -N- and -W-E-R) that are crucial and typical residues of the galectin family, related to carbohydrate-binding affinity (Fig. 3). Moreover, the main residues required for carbohydrate-binding of LhGal-1 were highly conserved with other galectins from fishes, mammals, birds, reptiles, and amphibians.
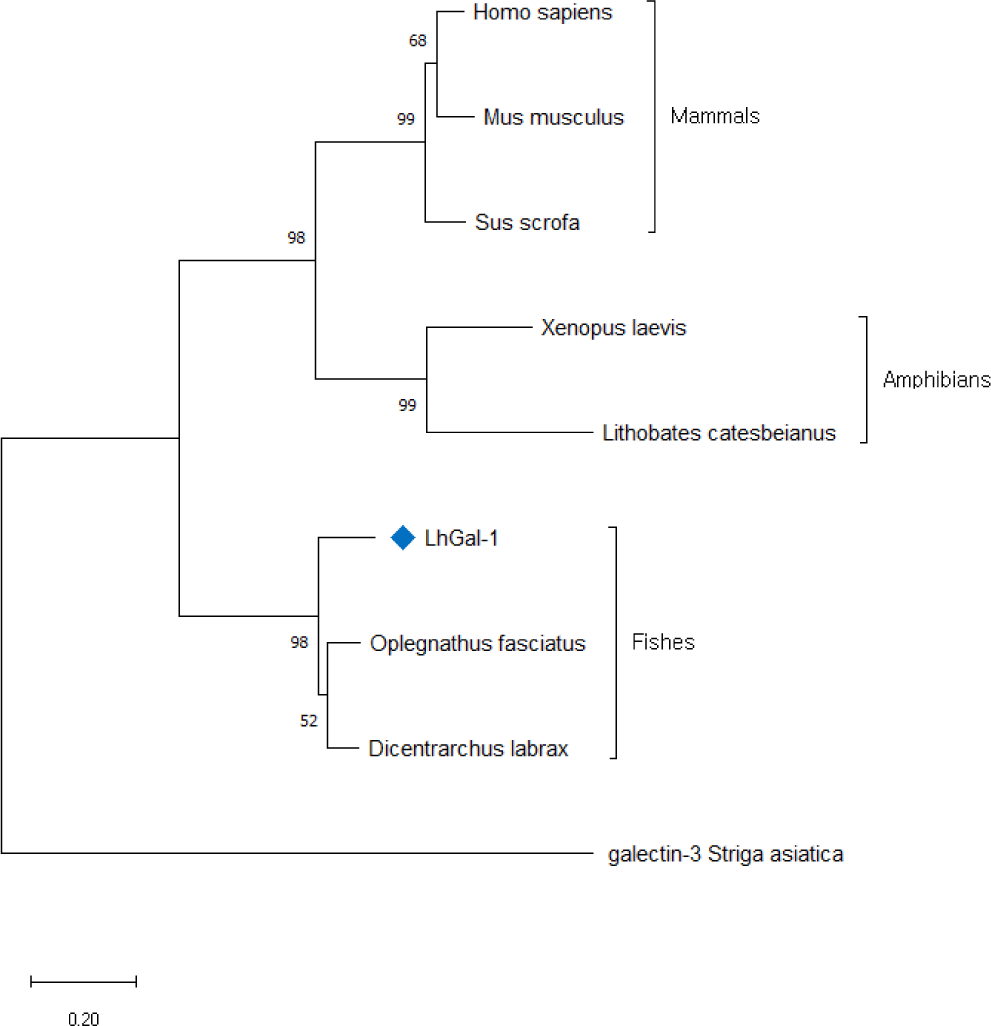
To comprehend the evolutionary relationship of redlip mullet LhGal-1, phylogenetic analysis was performed using the amino acid sequences for galectin-1 orthologs available in NCBI database (Fig. 4). The constructed phylogenetic tree was based on the neighbor-joining method with 5,000 bootstrap replicates. LhGal-1 clustered together with the fish species (Fig. 4). The output data of pairwise comparison and phylogenetic analysis suggested that LhGal-1 was closely related to galectin-1 from rock bream (Oplegnathus fasciatus) (Table 2).
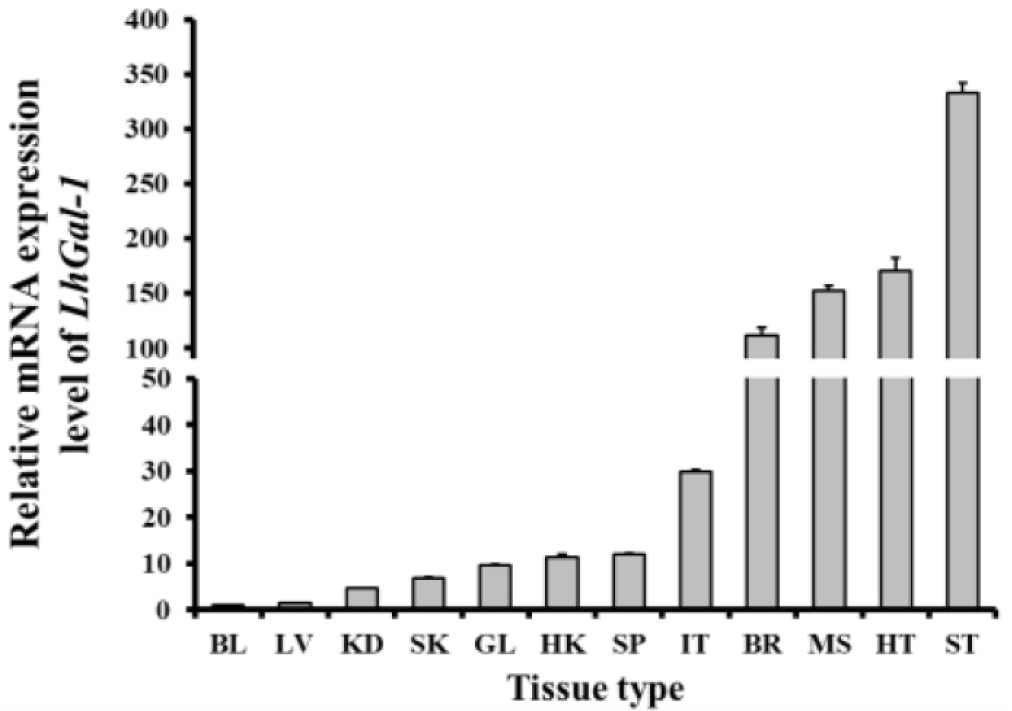
The mRNA analysis revealed that LhGal-1 was expressed in all the tested tissues. The stomach (333.20 folds) showed the highest mRNA expression level, followed by the heart (171.09 folds), muscle (152.25 folds), and brain (111.58 folds) (Fig. 5). The fundic, mucin, and epithelial cell surface glycocalyces are recognized by galectin-1 expressed in the gastrointestinal tract, which might be the reason for the highest expression of LhGal-1 in the stomach (Wasano & Hirakawa, 1997). Galectins are widely abundant in muscle tissues, some neurons, the thymus, and epithelial tissues. As per previous studies, Ctenopharyngodon idella and Epinephelus coioides galectin-1 are highly expressed in the muscle and heart (Chen et al., 2016; Zhu et al., 2019). Galectin-1 is involved in several biological processes as it interacts with multifarious receptors expressed in different cell types (Elola et al., 2005). Besides, galectin-1 binds to polylactosamine chains of laminin and promotes cell detachment and attachment in the extracellular matrix, which influences muscle development (Chen et al., 2016).
Time-dependent transcriptional regulation of LhGal-1 expression was examined in the gill and blood to estimate its dynamic expression profile in response to different immunogenic stimulants (Fig. 6). Fish gills are the main sites of pathogen entry and are a part of the first line of defense in aquatic organisms living in a pathogen-rich environment. Furthermore, blood is the pipeline for the migration of immune cells to the inflammation site. As immunomodulators of innate immunity, galectins are expressed in almost all immune cells and are involved in controlling related biological processes (Cerliani et al., 2011). Accordingly, gill and blood were selected to understand their complex functions following exposure to immune stimulants (LPS, typical cell wall components of gram-negative bacteria; poly I:C, structural analog of viral double-stranded RNA, L. garvieae, gram-positive bacteria). Following the injection of LPS and L. garvieae, LhGal-1 mRNA expression was significantly upregulated after 48 h in the gill (Fig. 6A). Consistently, the mRNA expression of galectin-1 in rock bream gill was reported to be significantly upregulated 48 h post injection of E. tarda (G–) and S. iniae (G+) (Thulasitha et al., 2016). The poly I:C-treated group showed significant upregulation between 48 and 72 h, but not between 6 and 24 h, in the gill (Fig. 6A). Similar early phase downregulation was reported in olive flounder (Paralichthys olivaceus) after injection of poly I:C (Liu et al., 2013). Moreover, expression of quadruple domain-containing galectin was downregulated in disk abalone until 12 h after injection of poly I:C and viral hemorrhagic septicemia virus (Gayashani Sandamalika & Lee, 2020). However, the reasons for early downregulation and possible underlying mechanisms remain unclear. It is possible that early phase downregulation of LhGal-1 in poly I:C-treated group helps in curbing the increase in the viral infection.
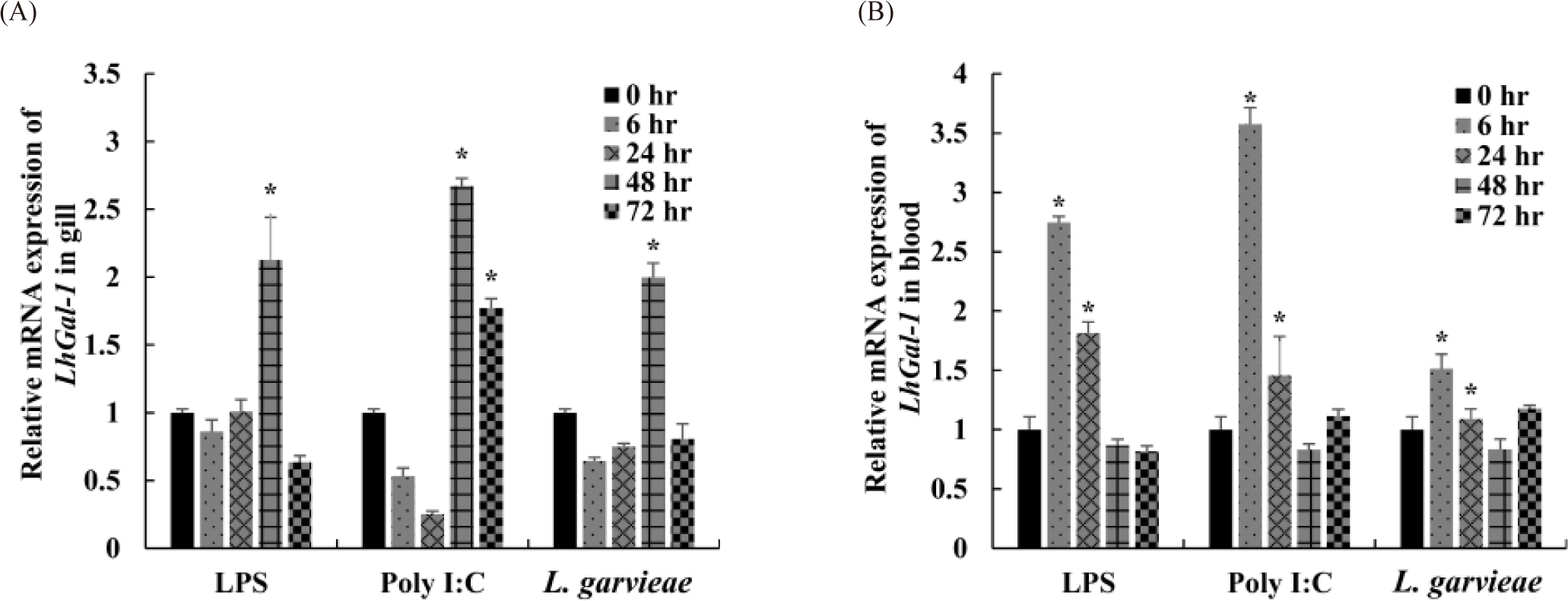
On the contrary, the relative LhGal-1 mRNA expression in blood was significantly upregulated 6 h post injection of LPS, poly I:C, and L. garvieae, and 24 h post injection of LPS and poly I:C (Fig. 6B). Consistently, early phase upregulation of galectin-1 was observed in mollusk (Solen grandis) hemocytes upon PAMP stimulation (LPS, peptidoglycans [PGN], and glucan) (Wei et al., 2012). Galectins expressed by immune cells not only recognize some pathogens as PRRs (PAMPs) but also contribute to innate immune cell activation and restore homeostasis against inflammation (damage-associated molecular patterns) (Sato et al., 2009). Taken together, these findings along with the previous studies suggested that in redlip mullet, LhGal-1 might act as a regulator of immune cells and modulate the activity of the encountered pathogen.
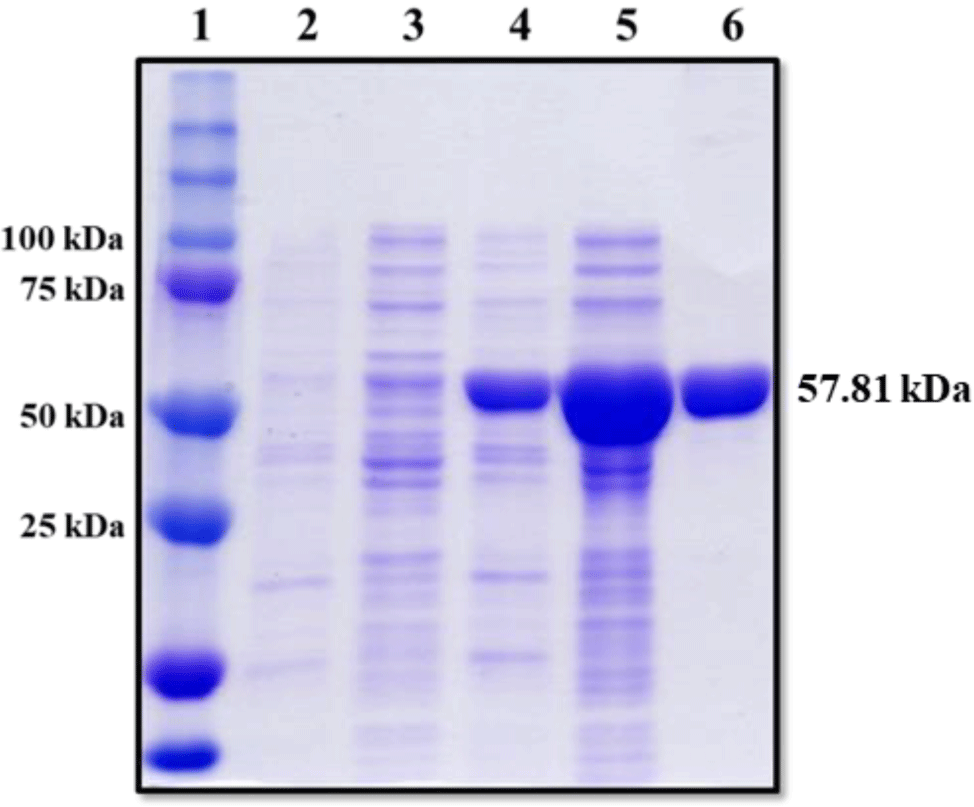
The coding sequence of LhGal-1 was cloned into a pMAL-c5X expression vector, and these plasmids were introduced into the E. coli strain, ER2523. The recombinant MBP (rMBP) fusion protein was obtained from IPTG-induced cells and purified using amylose affinity chromatography. SDS-PAGE was performed to visualize rLhGal-1 MBP fusion proteins and samples were obtained from each purification step; uninduced lysates, induced lysates, and eluted proteins. SDS-PAGE indicated no protein expression in the pellet and supernatant of the uninduced lysates, while specific bands were observed for the rLhGal-1-MBP fusion protein, at approximately 58 kDa (predicted molecular weights = rLhGal-1:15.31 kDa and MBP 42.5 kDa) (Fig. 7).
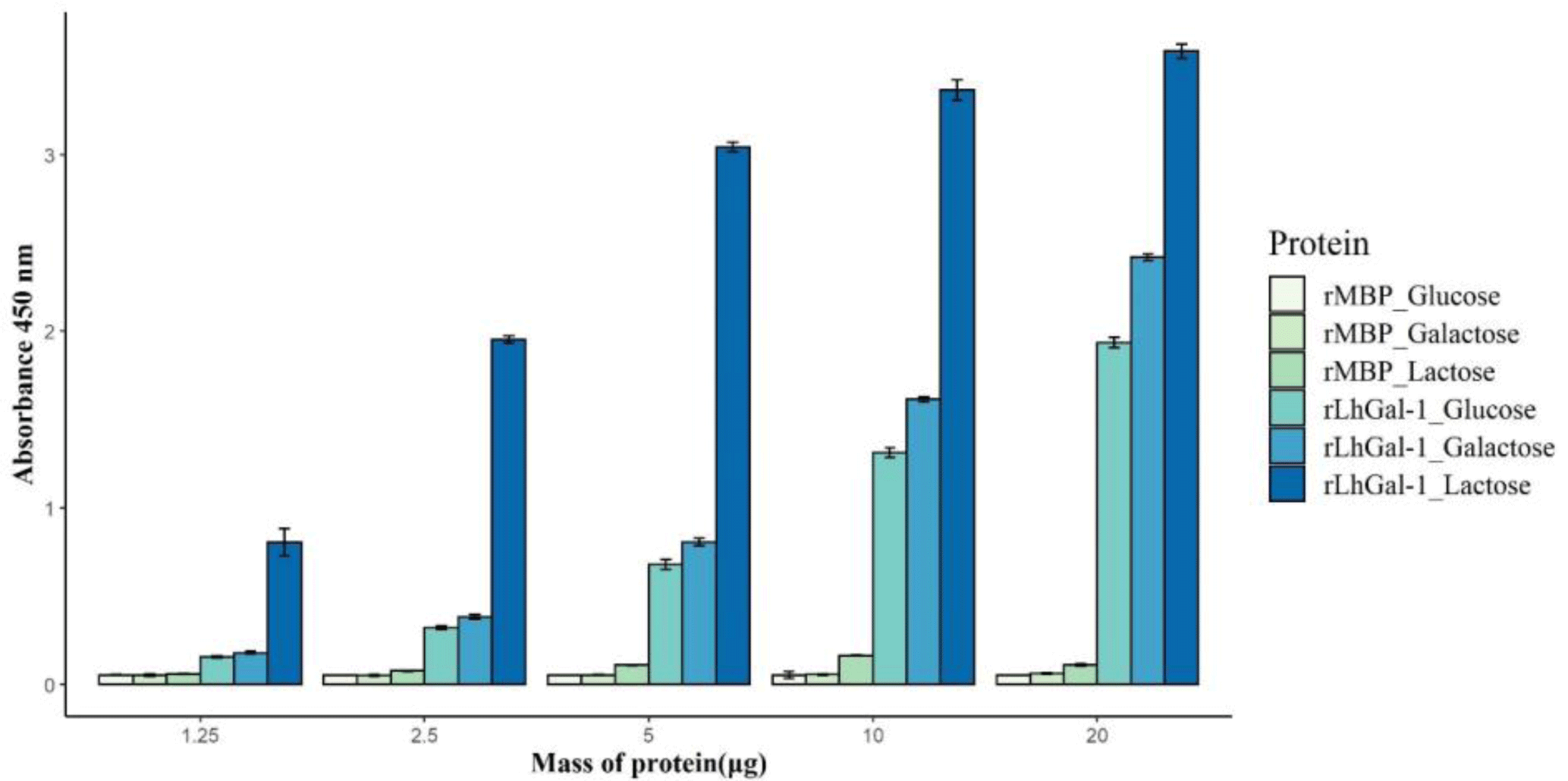
To investigate the carbohydrate specificity and binding ability of rLhGal-1, a sugar-binding assay was performed with three different carbohydrates as follows: α-lactose, D-galactose, and D-glucose. Although all carbohydrates bound to rLhGal-1, the disaccharide α-lactose showed a higher specificity than the monosaccharides galactose and glucose (Fig. 8). MBP-treated groups showed no significant activity with carbohydrates, which indicated that MBP might not have affected the binding properties of rLhGal-1 (Fig. 8). Significant differences noted in the binding ability of rLhGal-1 to lactose and galactose were possibly due to their structural features. LhGal-1 CRD forms a beta-sandwich with two skewed sheets and shapes the carbohydrate-binding site. Six strands in the beta-sheet form a groove that is long enough to capture linearized polysaccharides (Leffler et al., 2002). Although both galactose and lactose commonly possess beta-galactoside, rLhGal-1 has a higher affinity for lactose, which is longer than galactose. These results suggested that the binding ability of galectins is dependent on their affinity to the source as well as the length of their ligands.
The microbial binding ability of rLhGal-1 was evaluated using ELISA and revealed that rLhGal-1 could bind to all the tested G (+) and G (–) bacteria, whereas no binding activity was detected with the elution buffer and MBP controls (Fig. 9). A previous study on the binding activity of galectin from Eriocheir sinensis suggested that the interactions between rEsGal and PAMPs (LPS, PGN, and glycan) occur in a dose-dependent manner. Also, the highest binding activity was observed from 200 nmol/L of galectin toward LPS from E. coli (Wang et al., 2016). Among the PAMPs, LPS and PGN are major components of G (+) and G (–) bacterial cell walls, respectively. Thus, these results suggested that rLhGal-1 can recognize various pathogens and might act as a PRR involved in the innate immune response of the redlip mullet.
The microbial agglutination activity of rLhGal-1 was examined using E. coli. rLhGal-1 aggregated the tested bacteria while no agglutination was observed with rMBP (Fig. 10). The bacterial agglutination activity of rLhGal-1 further affirmed that LhGal-1 could be a PRR involved in the immune defense against bacteria. It might also serve as a facilitator of opsonization and phagocytosis by macrophages as shown for galectins in other systems (Cerliani et al., 2011).
Conclusion
We identified galectin-1 homolog from redlip mullet, named LhGal-1, and characterized its function. The in silico analysis was performed using several bioinformatics tools. To comprehend the potential role of LhGal-1 in the host immune system, the spatial and temporal mRNA expressions of LhGal-1 were estimated via qPCR. As a result, LhGal-1 was expressed in all tested tissues and upregulation was shown in gill and blood upon being challenged with immune stimulants. The carbohydrate-binding assay revealed that rLhGal-1 has a higher affinity to α-lactose than D-galactose and D-glucose. In addition, rLhGal-1 could bind to all tested bacteria and aggregate E. coli. Therefore, this study indicates that LhGal-1 may be an essential gene for recognizing invading pathogens and modulating host defense mechanisms.








