Introduction
The immunoglobulin superfamily (IgSF) is a large superfamily on the cell surface that is involved in the process of cell adhesion and recognition. Most of IgSFs are involved in the immune system and play roles such as antigen receptors, antigen-presenting molecules, and receptors on natural killer T-cells. IgSFs have hundreds of proteins with different functions, and immunoglobulin-like domains (Ig domains) can act as ligands between proteins (Williams & Barclay, 1988). The amino acid size of this domain is 70–110 amino acids (aa), classified according to size and function (Neil Barclay, 2003). The Ig domain has an Ig fold and is stabilized using strong disulfide bonds formed between hydrophobic amino acids and cysteines in the sandwich structure (Bork et al., 1994). One end of the Ig domain has a section called the complementarity determining region, which is essential for the specificity of the antibody for its ligand.
CD96 is a membrane-bound receptor for IgSF and was first discovered in humans in 1992 (Wang & Lincoln, 1993). CD96 is a single-pass transmembrane receptor with a heavily N-glycosylated site (Seth et al., 2007). The expression of CD96 is limited to natural killer (NK) cells and T cells and is closely related to CD155, the membrane-bound receptor of IgSF (Du Pasquier et al., 2004; Fuchs et al., 2004). In mammals, mice deficient in CD96 are more sensitive to lipopolysaccharide-induced toxins than wild-type mice due to an increase in interferon γ (IFNγ) by NK cells (Chan et al., 2014). In addition, studies on the treatment of tumours and the inhibition of metastasis using CD96 existed in mice (Blake et al., 2016; Brooks et al., 2018). In humans, CD96 has been downregulated in CD8 T cells of human immunodeficiency virus-1-infected patients, consistent with the assumption that it may be a potent producer of IFNγ and selectively inhibit T cell effector function, but more research is needed (Eriksson et al., 2012). In teleost, CD96 was characterized in Nile tilapia and was confirmed to have two Ig-like domains and an immunoreceptor tyrosine-based inhibitory motif (ITIM). In addition, it is mainly distributed in the cell membrane and cytoplasm, and it was confirmed that the interaction with Necl5 is also preserved (Xie et al., 2021). Studies on CD96 are active in mammals, but studies on CD96 in teleosts have not been reported without Nile tilapia.
Red seabream (P. major) is an important farmed fish species in Korea and Japan. With the development of aquaculture technology, large-scale farming is possible, but it is exposed to diseases due to excessive concentrations and environmental factors. It is especially susceptible to infection with red sea bream iridovirus (RSIV), so coping is important. However, there is a lack of basic research on immune genes to develop vaccines to cope. In this study, we obtained the cDNA sequence of PmCD96 from P. major and analyzed its molecular characteristics and mRNA expression patterns. As a result, we demonstrate the immunological capability of PmCD96.
Materials and Methods
Red seabream was received by the Gyeongsangnam-do Fisheries Resources Research Center (Tongyeong, Korea). Healthy red seabream selected using pathological clinical tests were incubated at 20°C for one week prior to use. All red seabream used in the experiment was euthanized using benzocaine (Sigma-Aldrich, St. Louis, MO, USA), and tissue was collected. Streptococcus iniae FP5228 and RSIV, the pathogens used in attack experiments to confirm pathogenicity, were obtained from the National Institute of Fisheries Sciences (Busan, Korea).
The open reading frame containing the domain of PmCD96 was identified in next-generation sequencing analysis using liver extracted from pathogen-infected red sea bream. The integrity of the cDNA sequence was confirmed by Sanger sequencing. The exact amino acid sequence and nucleotides of PmCD96 were analysed using the BLAST algorithm of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/blast). The specific domains of the amino acid sequence were predicted using the Simple Modular Architecture Research Tool (http://smart.embl-heidelberg.de/smart/set_mode.cgi?NORMAL=1), and the isoelectric point and molecular size were predicted using ExPASY ProtParam (https://web.expasy.org/protparam/). The N-glycosylated site was predicted using NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/services/NetNGlyc/). Multiple sequence alignment was performed using the ClustalW algorithm (https://www.ebi.ac.uk/Tools/msa/clustalo/). The phylogenetic tree was constructed using a neighbor-joining method with a bootstrap of 1,000 replicate datasets using Molecular Evolutionary Genetics Analysis version 10.0.
Total RNA was extracted from the trunk kidney, head kidney, liver, stomach, spleen, skin, muscle, intestine, eye, brain, heart, and gill of healthy red seabream using TRIzol reagent (Takara, Otsu, Japan). Total RNA extraction was performed according to the manufacturer’s instructions. The concentration and purity of the RNA samples were measured using a NanoVue spectrophotometer (GE Healthcare, Buckinghamshire, UK). cDNA synthesis was conducted using the PrimeScript 1st strand cDNA Synthesis Kit (Takara) according to the manufacturer’s instructions. Real-time quantitative real-time analysis was performed using 5-fold diluted cDNA as a template with TB GreenTM Premix Ex TaqTM (Takara) and a DICE Real-Time System Thermal Cycler (Takara). Relative mRNA expression was analyzed by the 2-ΔΔCT method (Livak & Schmittgen, 2001) and normalized to the EF-1α of red sea bream. Primer sequences are listed in Table 1.
| Primers | Sequence (5’-3’) | |
|---|---|---|
| PmCD96 | Forward | CAAGGACATGGAGAACATCT |
| Reverse | GGTTGCCATTTCACAAGTAT | |
| EF-1α | Forward | GGGAAAGGAAAAGATCCAC |
| Reverse | TTCTTCTGAGCCTTCTGTG | |
To confirm the expression pattern of PmCD96 in the host immune response after pathogen infection. Red seabream was divided into three groups according to each kind of pathogen: RSIV (1 × 106 copies/fish) or S. iniae (1 × 105 cells/fish) were intraperitoneally injected. During the experiment, the temperature of the seawater was maintained at 20 ± 1°C, and five fish were sampled from each group each time and day after infection (0 h, 1 h, 12 h, 1 d, 3 d, 5 d and 7 d). Then, the liver, spleen, head kidney, and gills were collected from each fish. Total RNA extraction, cDNA synthesis, and quantitative reverse transcription polymerase chain reaction were performed as described above.
Statistical analysis was performed using SPSS software version 19 (IBM, Armonk, NY, USA), and the expression of PmCD96 in tissues was assessed by one-way analysis of variance followed by Tukey’s honestly significant difference test. Experimental results are shown as ± SD, and all samples were analyzed in triplicate.
Results and Discussion
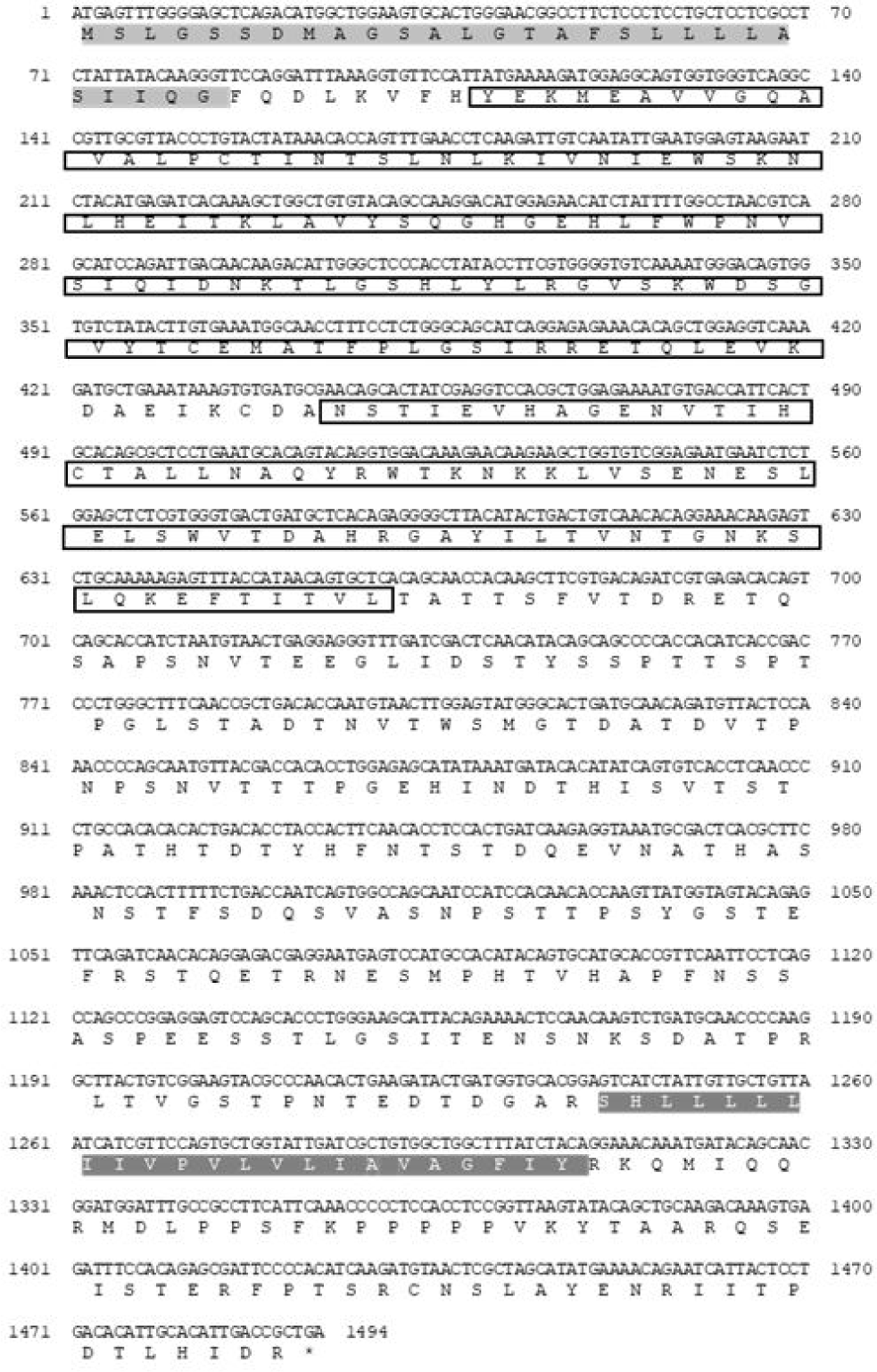
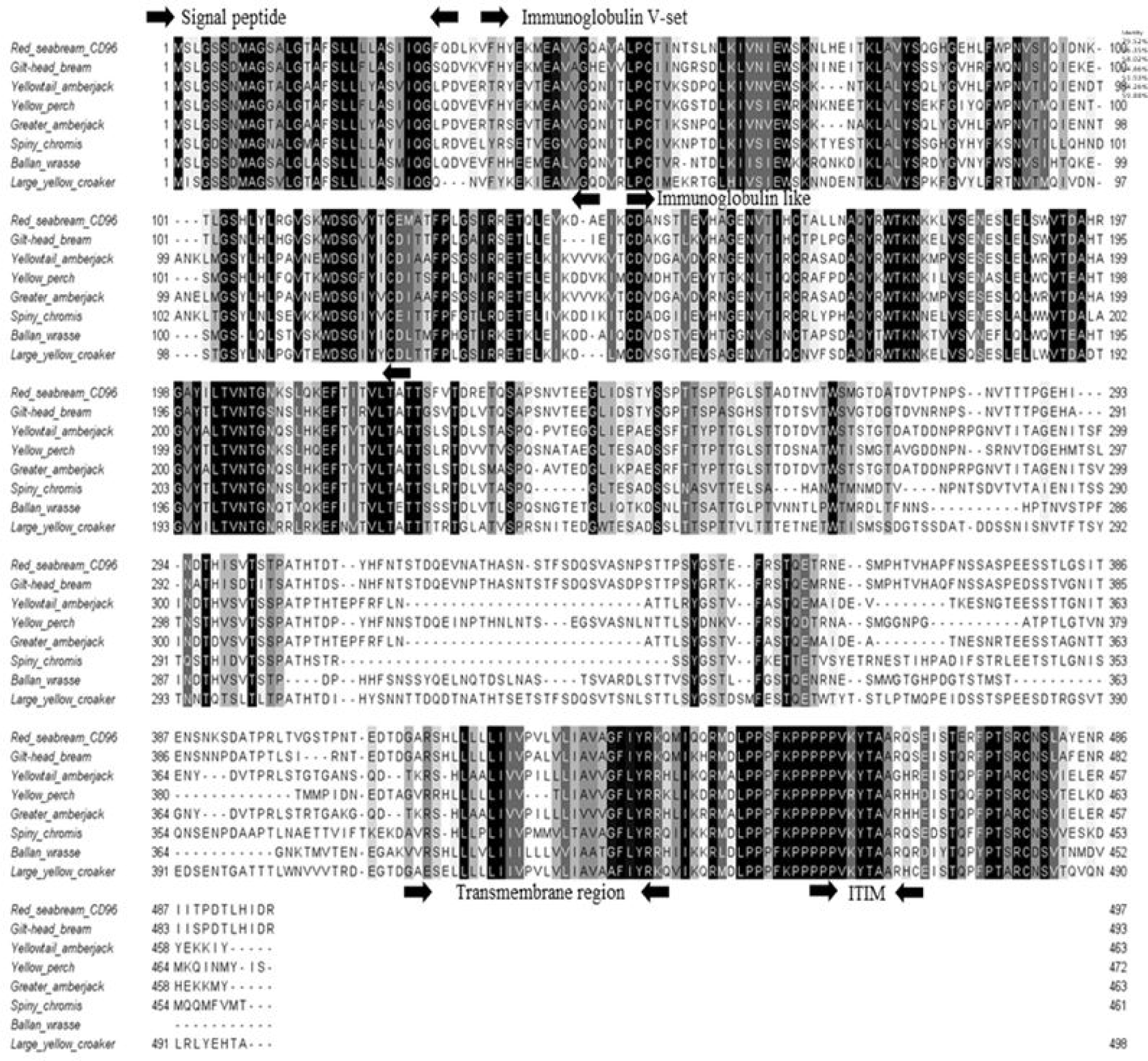
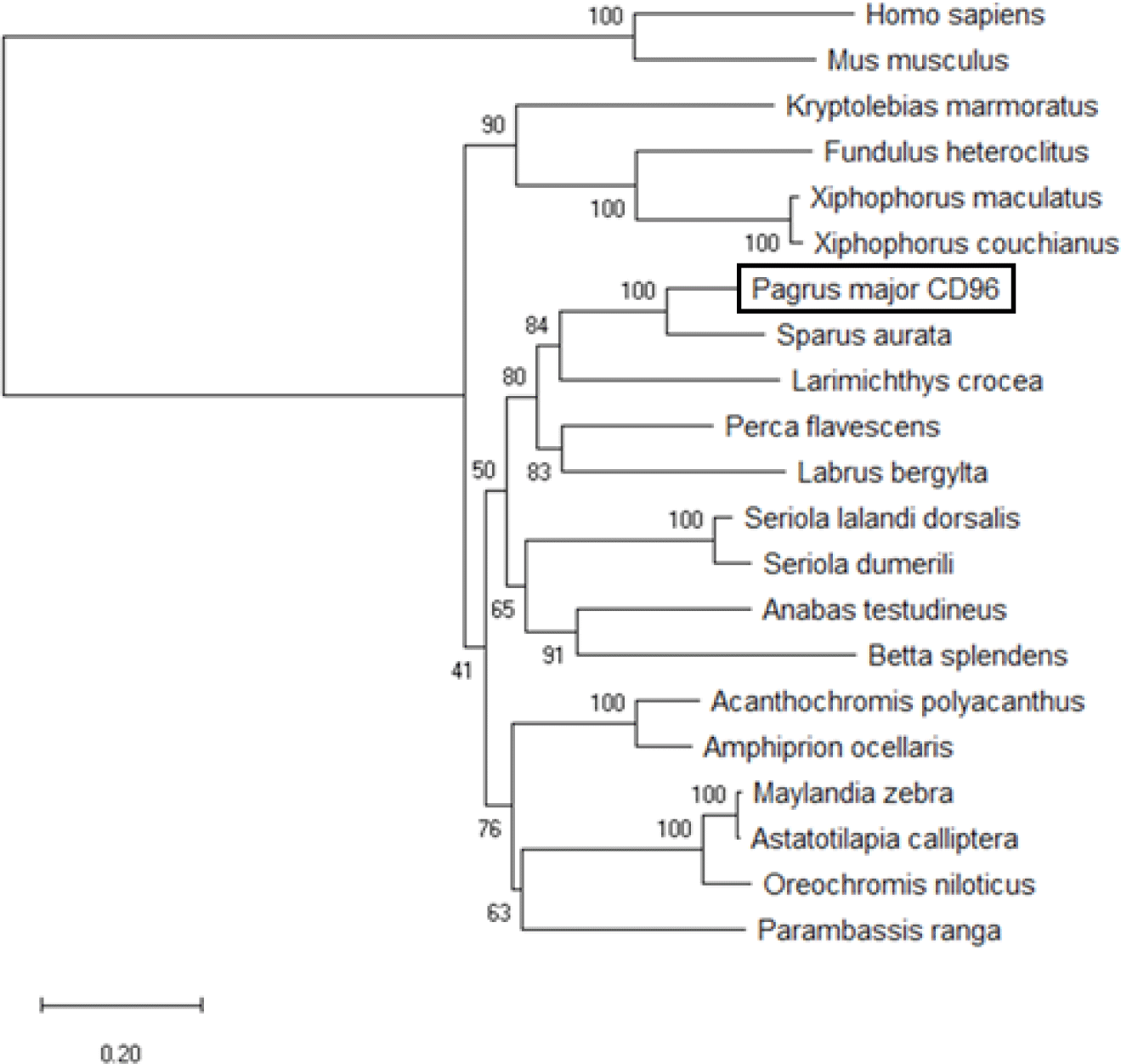
CD96 is a member of IgSF that shares a V-like domain with a transmembrane domain that is strongly N-glycosylated in mammals (Wang et al., 1992). In this study, the sequence of PmCD96 was obtained from red seabream, and its structure was confirmed. The PmCD96 sequence had a signal peptide (1–28 aa), two Ig domains (40–140 and 150–219 aa) and a transmembrane region (414–436 aa). The total amino acid content was 497 aa, the molecular weight was 54.2 kDa, with an isoelectric point of 5.43 (Fig. 1). In multiple sequence alignment analysis, identity was 51.93%–79.52%, and it was confirmed that the signal peptide, immunoglobulin, transmembrane region, and ITIM domain were less conserved (Fig. 2). Phylogenetic analysis using different CD96 genes showed that PmCD96 was in the same lineage as other fish, and PmCD96 was confirmed to be closely related to Sparus aurata (Fig. 3). The study of T cell immunity in fish is complicated by the fast development of genes related with T-cell immunity (Scapigliati et al., 2018). Due to these evolutionary constraints, T cell immune-related gene sequences may become less conserved across species. PmCD96 has just two lg-like domains structurally, compared to three in human CD96 and mouse CD96 (Georgiev et al., 2018). PmCD96 is binding location to the ligand requires more investigation. CD96’s cytoplasmic domain is short, but it contains several essential motifs, including ITIM and proline-rich motifs. An ITIM was found in the FcRIIB family of low-affinity immunoglobulin G (IgG) receptors. When co-aggregated with B-cell receptor by IgG immune complexes on the same cell, IgG receptors have long been known to decrease mouse B-cell activation, and this feature was considered to account for feedback regulation of antibody responses (Sinclair & Chan, 1971). The tyrosine in the receptor ITIM is phosphorylated by Src family protein kinases, which can engage protein tyrosine phosphatase and impede the transmission of the activation signal (Daëron et al., 2008). This can help to minimize excessive T-cell activation, which is beneficial for lowering inflammation. PmCD96 is sequence has intermediate homology with other species in general, but the essential functional domains are conserved, indicating that PmCD96 may have a comparable function to other CD96s.
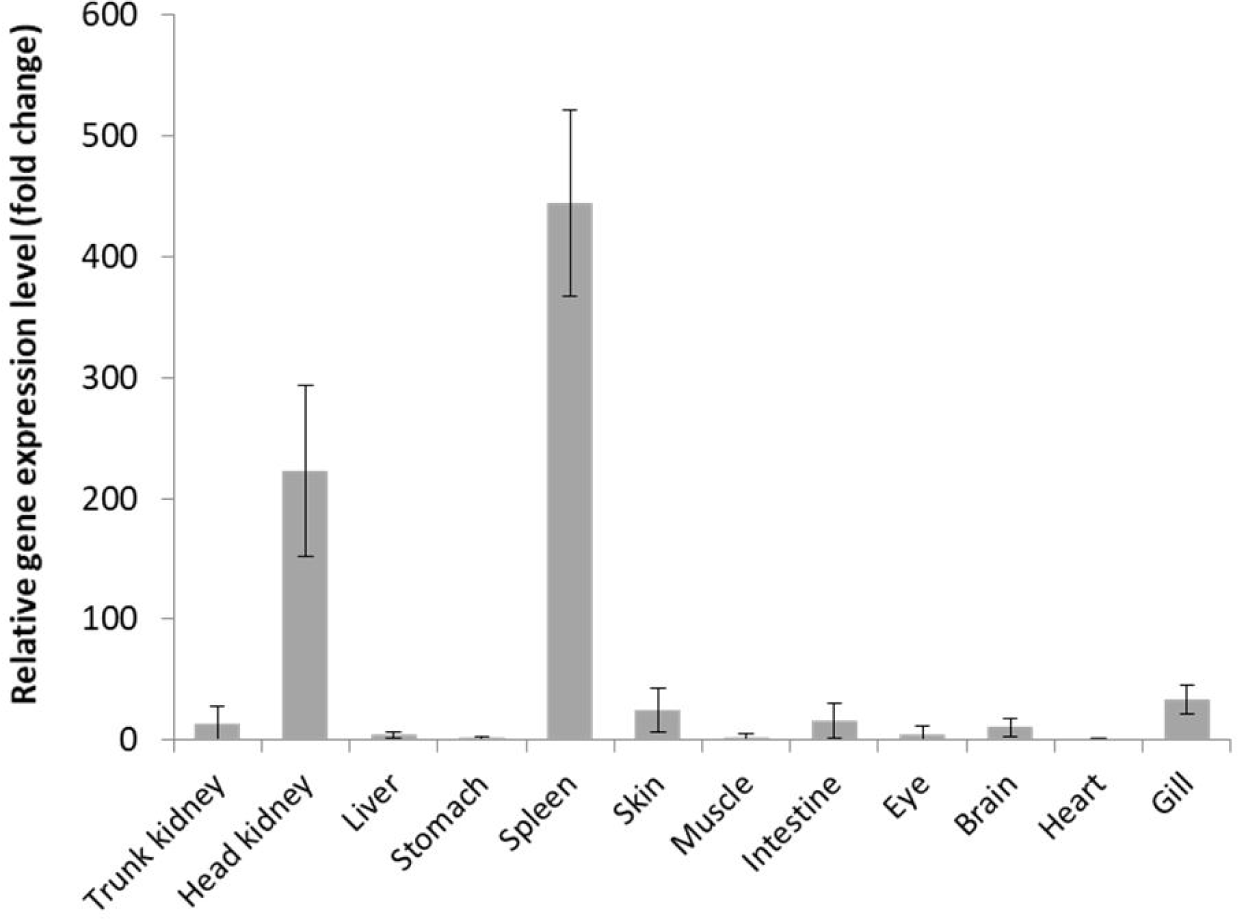
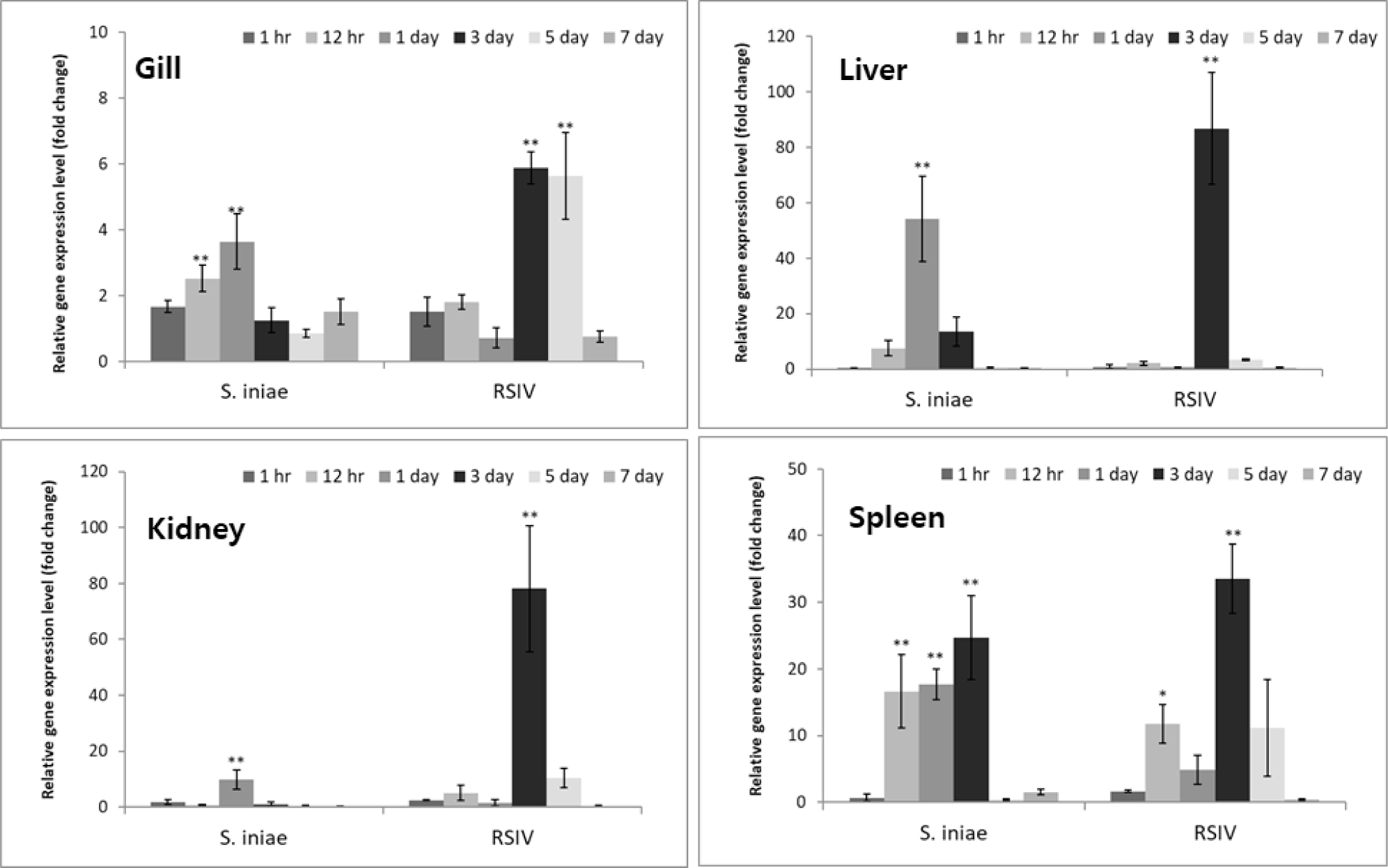
CD96 is known to be expressed mainly through T cell activation, and it has been shown to be primarily confined to T cells and NK cells, which are cells of haematopoietic origin (Fuchs et al., 2004; Wang et al., 1992). When the expression of normal red sea bream was confirmed in this study, a large amount was expressed in the head kidney and spleen (Fig. 4). These results, along with the results of studies confirming that the haematopoietic organs of fish are in the head kidney, and spleen, indicate that CD96 is also thought to be related to T cells and NK cells, which are also cells of haematopoietic origin (Pulsford et al., 1994). But in case of Nile tilapia the highest level of CD96 expression was found in the liver (Xie et al., 2021). These results suggest that further investigation into the normal function of CD96 in fish is needed. After red sea bream was infected with the pathogens S. iniae and RSIV, the expression of PmCD96 was analyzed in tissues known as immune organs. As a result, it was confirmed that all tissues infected with RSIV showed the highest expression value on the three-day post-infection (Fig. 5). Late after RSIV infection, PmCD96 showed a second surge of expression, which was most likely brought on by a delay in T cell-mediated acquired immunity (Xie et al., 2021). These results show that when compared with previous studies, the expression of immune genes appeared from the middle day post-infection (Hwang et al., 2014). After infection with S. iniae, PmCD96 showed the highest expression value on 1 dpi after infection in organs except for the spleen, which showed high expression on 5 dpi. S. iniae infected European seabass immunogene profile results confirmed that most of the immune genes used in the study were highest in the head kidney between 12 and 24 hours after infection (El Aamri et al., 2015). In mice, anti-T cell immunoreceptor with Ig and ITIM domains (TIGIT) monoclonal antibodies have exhibited remarkable antitumor activity in mouse tumor models when combined with anti-PD-L1 or anti-PD1. This mechanism is considered to entail TIGIT inhibitory role on regulatory T cells and CD8+ T cells (Johnston et al., 2014; Kurtulus et al., 2015). Looking at previous studies on the function of CD96 in mice and fish, these expression results show that CD96 of red sea bream functions as an immune gene. Also, CD96 is more sensitive to virus infection than bacteria, based on its expression pattern after infection in red sea bream. This could support the regulatory expression patterns discovered during RSIV infection in the gill, spleen, liver, and kidney. However, additional experiments, such as functional analysis, are needed to determine the exact mechanism of the PmCD96.
In conclusions this research by confirming the expression of CD96 in red seabream, the clear mechanism remains unknown, but its potential function as an immune gene appears to be confirmed and could be used as primary data in research on the immune system of fish.








