Introduction
The blue swimming crab (Portunus pelagicus) is a marine resource of significant economic importance for Thailand. Most blue swimming crab production comes from wild catches, as commercial farming has not yet been successful and involves high costs. In addition to being a popular seafood for domestic consumption, the blue swimming crab is a raw material in the processing and export sectors. Thailand produced 39,700 metric tons of blue swimming crab in 2023, valued at 8,453.1 million baht (Fishery Statistics Analysis and Research Group, 2024). The requirement to maintain a consistent capture of blue swimming crabs, combined with rising demand for consumption, processing, and export, has prompted fishermen to increase the efficiency of their fishing equipment. However, the natural blue swimming crab population has been gradually declining due to enhanced efficiency and the growth of the fishing crew (Onsri et al., 2024). To address these problems, the government has implemented blue swimming crab resource management, such as regulating fishing gear and protecting nursery areas for juvenile marine animals.
Over the past ten years, the crab bank project has emerged as a prominent example of this approach in Thailand. In crab hatcheries, the usual practice is to collect eggs from berried female crabs, hatch them into the zoea’s first larval stage, and then release them into the seawater (Thiammueang et al., 2012). For effective planning and management of crab bank projects, it is crucial to study the genetic information of the blue swimming crab. Interestingly, there has been limited research on the genetic information of blue swimming crabs for crab bank project management. Therefore, our study examined the genetic information of blue swimming crabs that enter the crab bank program to serve as a guideline for managing these crabs to maintain genetic diversity.
Genetic diversity refers to the variation in genetic makeup within a species, including diversity between populations and within a single population. This diversity is crucial for the survival of a species, as it enables organisms to adapt to changing environments and supports the potential for future evolution (Birader, 2023). Genetic structure is the pattern of genetic variation within and between populations caused by various factors (Gilleard & Redman, 2016). It involves characterizing populations based on gene frequency and examining changes in these frequencies caused by different factors. Studying the genetic structure helps to distinguish between subpopulations and identify individuals from mixed populations. Understanding the genetic structure of marine species is essential for formulating effective resource management strategies. Consequently, incorporating genetic data is vital to guiding and enhancing the management strategies of the crab bank project.
The mitochondrial DNA control region (mtDNA CR) exhibits a significantly higher mutation rate. Its nucleotide substitution rate is approximately 5–10 times greater than other regions in the mitochondrial genome and about 25–100 times higher than nuclear genes (Boore, 1999). Due to this characteristic, the mtDNA CR is particularly valuable for assessing genetic diversity. Studies on population genetic variation and structure in various marine crustacean species have utilized nucleotide sequences from this region. Examples include the black tiger shrimp (Penaeus monodon) (Wong et al., 2021), the blue swimming crab (P. pelagicus) (Suppapan et al., 2023), and the violet vinegar crab (Episesarma versicolor) (Supmee et al., 2012). Further, the haploid maternal inheritance of mitochondrial DNA facilitates the tracing of lineage transmission (Avise, 2000). Mitochondrial DNA sequencing also serves as a genetic marker for verifying parent-offspring relationships. Studies investigating the relationship between berried female crabs and their offspring using mitochondrial DNA sequencing include those on the swimming crab (Portunus trituberculatus) (Cai et al., 2020) and the blue swimming crab (P. pelagicus) (Suppapan et al., 2024). Our study employed mtDNA CR nucleotide sequencing to trace the progeny of berried female crabs released into the sea as part of the crab bank project.
More than 50% of the blue swimming crab production in the Gulf of Thailand comes from Bandon Bay, a significant blue swimming crab fishing area in Surat Thani Province, Thailand (Sawusdee et al., 2023). The Bandon Bay area’s blue swimming crab bank initiative has garnered support over the last ten years, and local fishermen have consistently cooperated with it. To conduct a preliminary investigation into the genetic diversity, genetic structure, and demographic history of blue swimming crabs, we selected three crab bank sites within Bandon Bay. The genetic data from this study aim to provide essential insights to support the effective management and conservation of crab bank initiatives in the region. Additionally, this study analyzed the similarity of haplotypes between berried females from the crab bank project and wild crab populations after the release of offspring, aiming to assess the effectiveness of these releases over time. The findings from this research provide valuable insights for conserving the genetic diversity of blue swimming crab populations in Bandon Bay.
MATERIALS AND METHODS
Under approval number IAC 02-01-2024, the Rajamangala University of Technology Srivijaya Institutional Animal Care and Use Committee authorized the protocols used in this investigation.
Samples of berried female crabs were collected around three crab banks (Had Somboon crab bank, Tachana District; Ban Laem Poh crab bank, Chaiya District; and Ban Nangkam crab bank, Donsak District, Surat Thani Province), covering an area of 10 km2, using gill nets in early February 2024 (Fig. 1). Berried females were raised in tanks to allow them to release their eggs. After the eggs hatched into the zoea stage, they were released into the sea around the crab banks, covering a 10 km2 area. The release of crab larvae was carried out from late February to early March (Table 1).
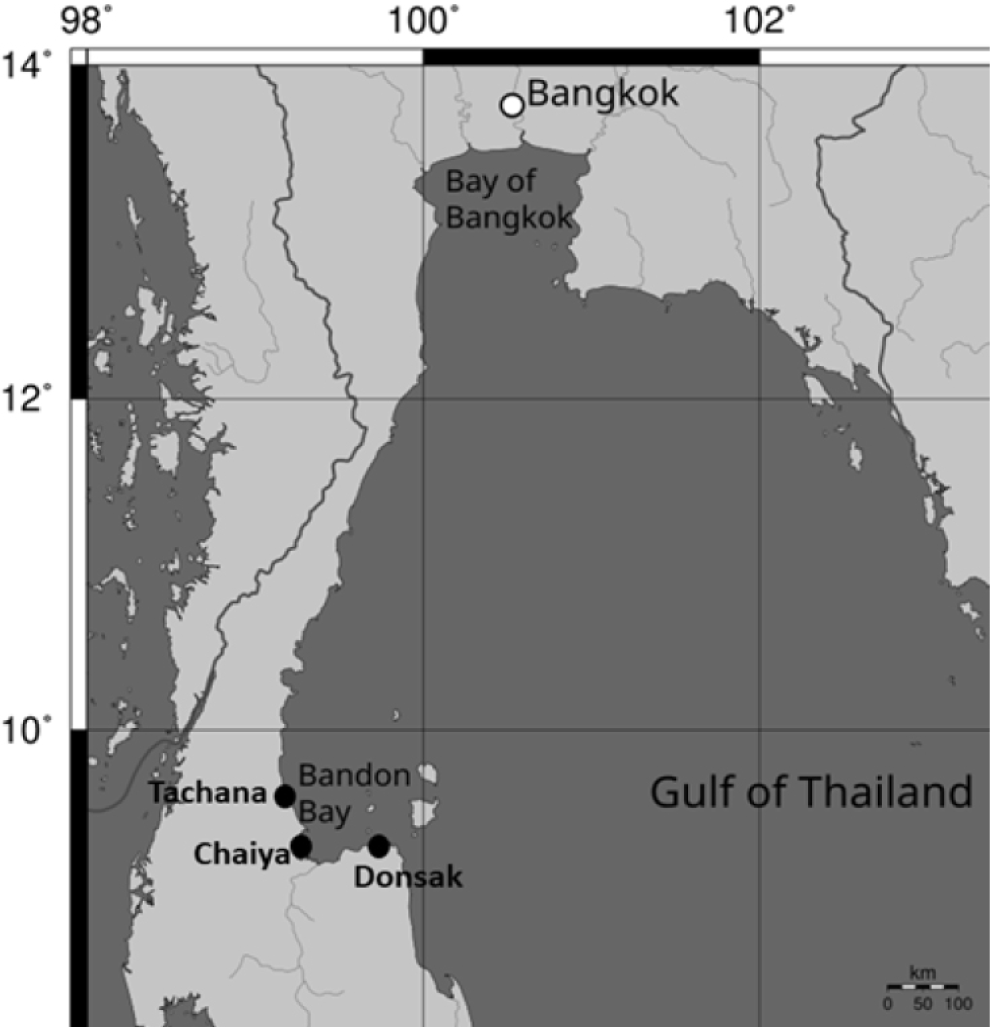
About five months after the release of the crab larvae (from 22 June–20 July 2024), a random sampling of 100 wild crabs with carapace sizes ranging from 8 to 15 cm was conducted in the area around the crab banks, covering 10 km2 (Fig. 1, Table 2). The single walking legs of berried female and wild crabs were clipped and preserved in 95% ethanol for transport to the laboratory and stored at a temperature of –20°C for subsequent DNA extraction. Genomic DNA was extracted from the muscle tissue of the walking legs using a commercial extraction kit (TIANGEN, Beijing, China) following the manufacturer’s instructions.
We designed a pair of primers to amplify the target mtDNA CR (600 bp): PUMA_CR_F (5’-TTG AAA TTA AAT CGA TTA TAT AG-3’) and PUMA_CR_R (5’-CAA GAG AAA AGG AAG AAA G-3’). A 50 µL PCR reaction mixture comprising 5 µL 10X Taq buffer, 5 µL 50 mM MgCl2, 4 µL 2 mM dNTP mix, 2 µL 10 µM forward primer, 2 µL 10 µM reverse primer, 0.5 µL Taq DNA polymerase (5 units, Bio-Helix, New Taipei, Taiwan), 5 µL DNA template (50–100 ng), and 26.5 µL ultrapure water. The amplification was operated on a thermocycler (Major Cycler, CYCLER, Taoyuan, Taiwan). PCR conditions consisted of 1 cycle of initial denaturation at 94°C for 4 min, 35 cycles of denaturation at 94°C for 40 sec, annealing at 45.7°C for 1 min, and extension at 72°C for 1 min, and one cycle of final extension at 72°C for 10 min. PCR product size was verified by 1% agarose gel electrophoresis. Amplified products were purified using the Gel/PCR Purification Mini Kit (TIANGEN) according to the manufacturer’s instructions and sent to nucleotide sequencing at ATGC, Thailand.
The nucleotide sequences obtained from the service unit were validated using the BioEdit Sequence Alignment Editor (Hall, 1999). The sequences were then analyzed to confirm their location within the mtDNA CR by referencing the NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Sequences were aligned using ClustalW version 1.83 (Thompson et al., 1994) and subsequently edited. Genetic diversity was assessed by calculating nucleotide diversity (π), haplotype diversity (h), and the mean number of nucleotide differences among all haplotypes using DnaSP version 6.00 (Rozas et al., 2017). Additionally, the presence of shared and unique haplotypes between berried females and wild crabs was examined. The exclusion of mtDNA CR was determined using the formula: the count of wild crabs with unique haplotypes divided by the total number of wild crabs.
Population genetic structure was analyzed using analysis of molecular variance (AMOVA) to compare the levels of genetic diversity within and between populations by examining F-statistic (ΦST) values. Genetic differentiation between populations was assessed by calculating pairwise FST values. All analyses were conducted with 10,000 permutations using the ARLEQUIN program version 3.5.1.2 (Excoffier & Lischer, 2010). Genetic distance (D) among populations was analyzed using the γST parameter with the DnaSP program version 6.00 (Rozas et al., 2017). Gene flow (Nm) between populations was estimated from pairwise FST values using the formula Nm = (1 – FST) / 2FST (Slatkin, 1987). A phylogenetic tree was constructed from the nucleotide sequences of each sample using the Neighbor-Joining method with the Kimura 2-parameter model and 1,000 bootstrap, using MEGA version 7 (Kumar et al., 2016). A haplotype network (MSN) was constructed based on the mean pairwise differences between haplotypes, calculated using the ARLEQUIN program version 3.5.1.2 (Excoffier & Lischer, 2010) with 10,000 permutations. The network was then manually drawn.
Three methods were employed to examine the demographic history of the blue swimming crab population. First, we conducted selective neutrality tests by analyzing Tajima’s D (Tajima, 1989) and Fu’s Fs (Fu, 1997) to evaluate any deviations from neutral evolution. Second, we performed a mismatch distribution analysis under the sudden expansion model to explore evidence of population expansion. Harpending’s Raggedness Index was employed to assess the compatibility of the observed data with the model, and the sum of squared deviations (SSD) was used to evaluate the goodness-of-fit. Third, we estimated the population size using the parameters θ0 and θ0, which represent 2Nμ, where N is the effective female population size. All analyses were performed with 10,000 permutations using ARLEQUIN software version 3.5.1.2 (Excoffier & Lischer, 2010).
RESULTS
The mtDNA CR sequence ranged from 391 to 396 base pairs. In the alignment of berried female crabs, 397 aligned sites were identified, comprising 13 gaps or missing data, 234 monomorphic sites, and 150 polymorphic sites. Among the polymorphic sites, 46 were singletons, and 104 were parsimonious-informative sites, resulting in 204 haplotypes. Of these shared haplotypes were 22, five were shared between populations, and 17 within a population, while 182 were unique (Fig. 2A). Haplotype diversity values for berried females ranged from 0.965 to 0.981, with nucleotide diversity varying between 0.020 and 0.052. Overall, the haplotype and nucleotide diversity across all berried female populations were 0.980 and 0.038, respectively (Table 3). In wild crabs, the alignment revealed 397 aligned sites, including 14 gaps or missing data, 198 monomorphic sites, and 185 polymorphic sites. These polymorphic sites consisted of 60 singleton sites and 125 parsimonious-informative sites. A total of 251 haplotypes were identified, of which 34 were shared: 23 between populations and 11 within a population. The remaining 217 haplotypes were unique (Fig. 2B). Haplotype diversity values for wild crabs ranged from 0.994 to 0.999, with nucleotide diversity spanning from 0.027 to 0.056. Overall haplotype and nucleotide diversity for all wild crab populations were 0.997 and 0.042, respectively (Table 3). A summary of the genetic diversity parameters for berried female and wild crab populations in Bandon Bay is provided in Table 3.

For the Tachana population, 130 haplotypes were identified among 114 berried females and 119 wild crabs. Seventy-seven unique haplotypes observed in 32 berried females or 45 wild crabs were excluded. The remaining dataset comprised 82 berried females and 74 wild crabs, which shared 53 haplotypes (Table 4). In Chaiya, 130 haplotypes were identified among 103 berried females and 105 wild crabs. After excluding 70 unique haplotypes, we found 27 berried females and 43 wild individuals. The analysis focused on 76 berried females and 62 wild crabs, which shared 60 haplotypes (Table 4). Similarly, in Donsak, 124 haplotypes were detected among 107 berried female crabs and 110 wild crabs. A total of 85 unique haplotypes, 35 berried females and 51 wild crabs, were removed. The final analysis included 72 berried females and 59 wild crabs, which shared 39 haplotypes (Table 4). In total population, 41.61% of wild individuals were excluded from the analysis (Table 4).
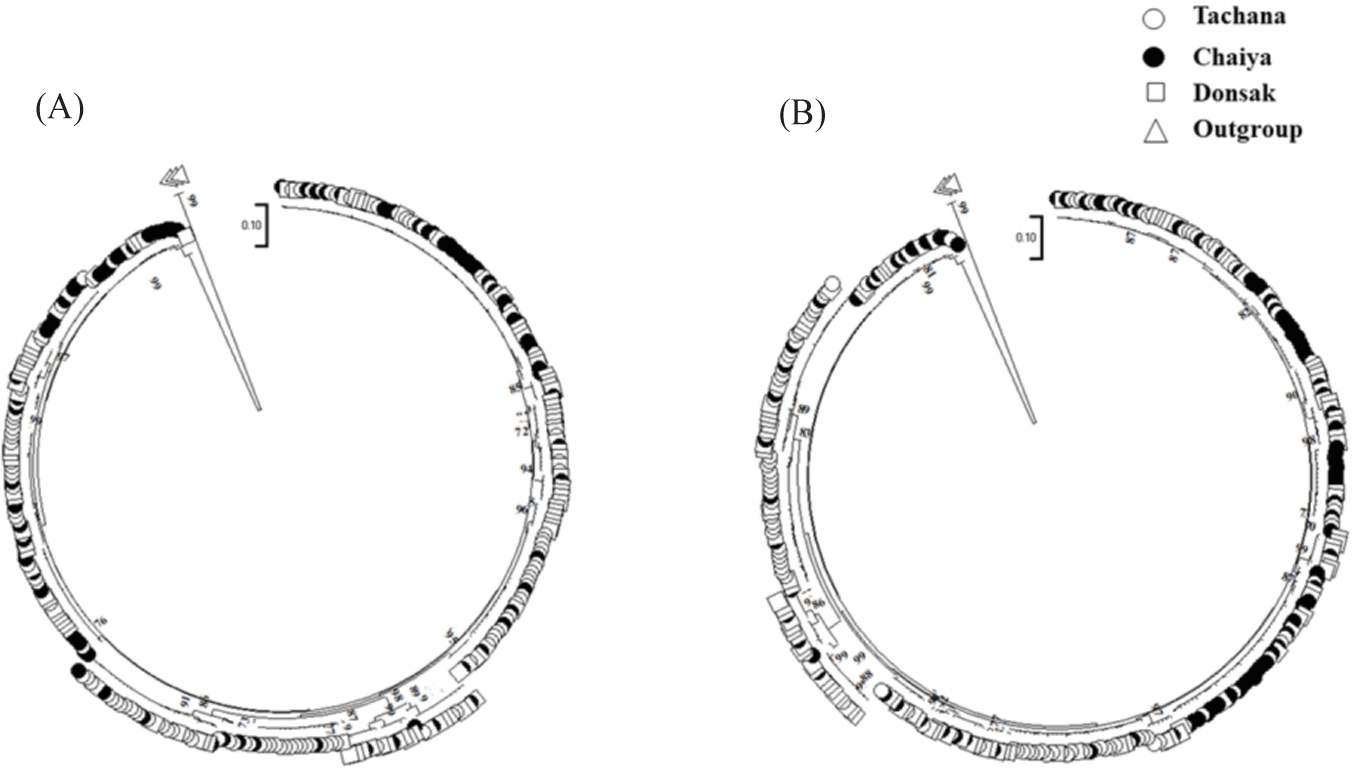
The genetic structure of berried female and wild blue swimming crab populations in Bandon Bay was analyzed. Molecular variance analysis indicated no significant population differentiation among berried female crabs across the three locations, as evidenced by the ΦST statistic (ΦST = 0.15500, p = 0.098). Similarly, the analysis for wild crabs revealed no distinct genetic structure among populations in Bandon Bay, with a ΦST value of 0.11547 (p = 0.089) (Table 5). Comparisons of the genetic structure between berried females and wild crabs in each location also showed no significant differences. In Tachana, the ΦST value was –0.00478 (p = 0.78020). Similarly, no significant differences were observed in Chaiya (ΦST = 0.00028, p = 0.34842) or Donsak (ΦST = –0.00503, p = 0.91149) (Table 5). These results indicate a lack of genetic differentiation between berried female and wild crab populations in Bandon Bay. Pairwise FST analysis revealed no significant differences among the berried female populations in Tachana, Chaiya, and Donsak, indicating an absence of population structure among berried female crabs in Bandon Bay (Table 6). Similarly, no significant genetic variations were detected among wild crab populations in Bandon Bay, confirming a lack of genetic structure in these regions (Table 6). The pairwise FST analysis between berried females and wild crabs in Tachana also showed no significant genetic differences (FST = –0.00478, p = 0.77705). Likewise, no significant differences were observed between berried females and wild crabs in Chaiya (FST = 0.00028, p = 0.35294) or Donsak (FST= –0.00503, p = 0.91031). The genetic distances among berried female crab populations were 0.13381 between Tachana and Chaiya, 0.07409 between Tachana and Donsak, and 0.06796 between Chaiya and Donsak. Similarly, in wild crab populations, the genetic distances were 0.09344 between Tachana and Chaiya, 0.05537 between Tachana and Donsak, and 0.05409 between Chaiya and Donsak (Table 7). The gene flow (Nm) values for berried female crabs were 1.679789 between Tachana and Chaiya, 3.340246 between Tachana and Donsak, and 3.704861 between Chaiya and Donsak. The Nm values of wild crab were 2.549152 between Tachana and Chaiya, 4.645092 between Tachana and Donsak, and 4.805040 between Chaiya and Donsak (Table 8). The phylogenetic analysis showed no distinct lineages among berried female crab populations in Bandon Bay (Fig. 3A). Similarly, the phylogenetic tree indicated that the wild crab population in Bandon Bay did not form any separate groups (Fig. 3B). The haplotype network of berried female crabs exhibited a complex structure, lacking a clear phylogeographic pattern among the 204 haplotypes. The most common haplotype (H7) was shared across three regions, forming a star-like network and directly connecting to other haplotypes from these regions in a short genealogical arrangement. This haplotype was present in 38 berried female crabs out of 324, representing 11.72% of the total (Fig. 2A). Similarly, the haplotype network of wild crabs displayed a high level of complexity, with no distinct phylogeographic structure among the 251 haplotypes. Haplotype H44, the most common among wild crabs, also formed a star-like network and was linked to other haplotypes in an extended genealogical tree. This haplotype was found in 6 wild crabs out of 334, accounting for 1.79% (Fig. 2B).
| Population | Tachana | Chaiya | Donsak |
|---|---|---|---|
| Tachana | - | 0.16398 | 0.09718 |
| Chaiya | 0.22938 | - | 0.09425 |
| Donsak | 0.13020 | 0.11891 | - |
| Population | Tachana | Chaiya | Donsak |
|---|---|---|---|
| Tachana | - | 0.09344 | 0.05537 |
| Chaiya | 0.13381 | - | 0.05409 |
| Donsak | 0.07409 | 0.06796 | - |
| Population | Tachana | Chaiya | Donsak |
|---|---|---|---|
| Tachana | - | 2.549152 | 4.645092 |
| Chaiya | 1.679789 | - | 4.805040 |
| Donsak | 3.340246 | 3.704861 | - |
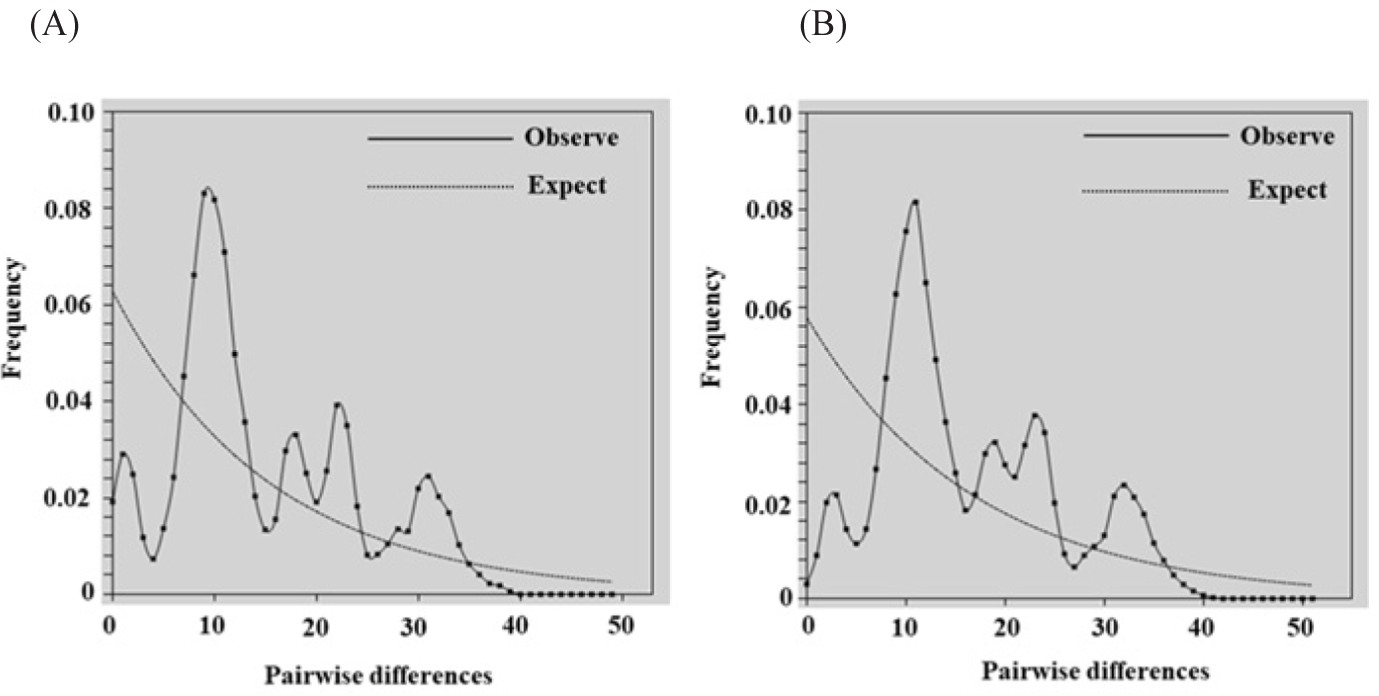
The demographic history of berried female crabs was assessed using three methods. First, Tajima’s D was –1.11418, and Fu’s Fs was –23.67344, showing statistically significant differences (Table 9). Second, the mismatch distribution was multimodal (Fig. 4A). The Rag index values (0.00381) were not statistically significant (Table 9). The pooled population‘s mismatch distribution did not fit a sudden expansion model, as indicated by the measured SSD from the goodness-of-fit test (SSD = 0.00644, p = 0.005) (Table 9). Finally, across all sampling sites, θ1 was greater than θ0, with values of 9.55195 for θ0 and 73.67188 for θ1 (Table 9). The demographic trajectory of wild crabs was analyzed. Tajima’s D was –1.33353, and Fu’s Fs was –23.61756, indicating statistical significance (Table 9). The mismatch distribution was also multimodal (Fig. 4B). The Rag index values (0.00279) were not statistically significant (Table 9). The observed SSD from the goodness-of-fit test (SSD = 0.00471, p = 0.007) showed that the mismatch distribution of the pooled population did not fit a sudden expansion model (Table 9). Finally, θ1 exceeded θ0 across all locations, with θ0 at 10.07578 and θ1 at 146.79688 (Table 9).
DISCUSSION
Our study revealed many distinct mtDNA CR haplotypes in berried females and wild crabs. Multiple unique haplotypes highlight the substantial effective female population size of the blue swimming crab in Bandon Bay. The effective female population size reflects successful female reproduction. The high number of effective female blue swimming crabs lowers the risk of inbreeding, potentially facilitating faster population recovery. Therefore, this highlights the ongoing significance of continued conservation efforts. The success of the artificial propagation program for blue swimming crabs depends on maintaining genetic diversity. Thus, the substantial effective female population size suggests a favorable prospect for the future recovery in Bandon Bay.
Samples from berried females and wild crabs exhibited high haplotype variation but relatively low nucleotide diversity. This pattern suggests a recent population expansion in the blue swimming crab. In rapidly expanding populations, genetic traits are shaped by new mutation accumulation and the preservation of high haplotype diversity (Watterson, 1984). Similar patterns of high haplotype diversity coupled with low nucleotide diversity have been reported in various marine crabs, including the horseshoe crabs (Tachypleus gigas) (Aini et al., 2021) and freshwater mitten crab (Eriocheir japonica) (Oh et al., 2023).
Nucleotide diversity, which measures polymorphism within a population (Nei & Li, 1979), was 0.038 in berried females and 0.042 in wild crabs. These values suggest that the nucleotide diversity of the blue swimming crab population in Bandon Bay is comparable to that in the Andaman Sea of Thailand (0.042) (Suppapan et al., 2023). Furthermore, the nucleotide diversity of blue swimming crabs in Bandon Bay is higher than that of other crustacean species. For example, nucleotide diversities in the swimming crab (Portunus sanguinolentus) (0.011) (Lu et al., 2022) and the swimming crab (Portunus trituberculatus) (0.020) (Cho et al., 2009) were all lower. Additionally, our data show that nucleotide diversity was higher in wild crabs than in berried females, indicating that wild crabs possess higher genetic diversity than berried females.
Although mtDNA CR, known for its high genetic diversity (Avise et al., 1987), can be used to determine the genetic relationships between individuals, it is specifically applied in stock enhancement programs to identify the parentage of offspring. The studies have employed it as a complementary tool to gain further insights, particularly in the case of the swimming crab (Portunus trituberculatus) (Cai et al., 2020). Given the substantial nucleotide variation in mtDNA CR fragments, it can be expected to perform well, especially in species with high haplotype diversity (Cai et al., 2020). In this study, an initial screening was conducted using mtDNA CR fragments. Considering its cost-effectiveness and efficiency, using mtDNA CR fragments appears to be a highly effective approach for the initial screening phase in future stock enhancement program evaluations.
In this study, common haplotypes were found in small proportions among the samples, accounting for 11.72% in berried female crabs and 1.79% in wild crabs. Furthermore, we identified a significant number of unique haplotypes. Consequently, the influence of unreleased crabs on haplotype distribution is considered negligible. In our study, 139 individuals of wild crabs (41.61%) were excluded due to the absence of a shared haplotype with berried females. As a result, the haplotype similarity between berried females from the crab bank project and wild crabs was 58.39%. These findings indicate that more than half of the wild blue swimming crabs share haplotypes with broodstock crabs, suggesting that over half of the wild crabs caught may have originated from the crab bank project. This study yielded results consistent with previous investigations on haplotype sharing between berried female blue swimming crabs (P. pelagicus) and wild crabs from crab bank projects in Trang and Krabi, Thailand (Suppapan et al., 2024), as well as similar findings in swimming crabs (Portunus trituberculatus) in Shandong Province, China (Cai et al., 2020). Both studies reported that more than half of the haplotypes shared between berried females and wild crabs originated from crab bank projects.
Our study suggests that the crab bank project implementation in Bandon Bay is supporting the recolonization of the blue swimming crab population. The findings indicate that releasing crab offspring into the wild contributes to the return of adult crabs, potentially boosting the wild population to a level that supports sustainable fishing practices. However, as this study is preliminary, further research is needed to validate these results by sampling crabs from areas beyond Bandon Bay. Expanding the analysis of mtDNA CR sequences, as performed in this study, could offer insights. Consequently, incorporating other genetic markers from the nuclear genome in future studies is recommended.
An analysis of the genetic structure of berried crabs in Bandon Bay, using six different methods, found no significant genetic differentiation within the berried crab populations in the area. Similarly, no population structure differences were observed among wild crabs associated with the crab banks. Our finding demonstrated that population genetic structure analyses indicated the absence of distinct genetic structuring in blue swimming crab populations within Bandon Bay.
Planktonic larvae mixing through water circulation promotes the genetic homogeneity of the blue swimming crab population. Bandon Bay is a shallow bay with an average depth of approximately 3 meters. Daily tidal changes are the main factor influencing the bay’s water circulation and tidal current patterns. Currents flow into the bay at high tide and out of the bay at low tide. Additionally, the bay’s water circulation varies seasonally. Currents move inside and outside the bay between October and February due to the northeast monsoon. The southwest monsoon pushes currents out of the bay from May to September. During the changeover period in March and April, present patterns change correspondingly. Furthermore, freshwater input from the Tapi River and other tributaries into the bay influences water flow direction (Jutagate & Sawusdee, 2022). These combined factors contribute to significant water circulation within Bandon Bay (Wattayakorn et al., 2001), facilitating gene flow throughout the bay.
Furthermore, the extended duration of the planktonic stage enhances population mixing, promoting genetic connectivity. The larval phase of blue swimming crabs lasts approximately 26–45 days, allowing for extensive dispersal (Efrizal, 2016). Many marine species exhibit life stages where gametes, larvae, or adults are free-moving in the open ocean, which promotes genetic connectivity (Uthicke & Benzie, 2003). Notably, marine species with longer larval durations often exhibit higher levels of genetic variation within populations (Russo et al., 1994). The extended planktonic larval stage of blue swimming crabs enhances opportunities for gene flow, as observed in other marine crabs with similar life-history traits, such as shore crab (Carcinus maenas) (40 days) (Moksnes et al., 2014) and helmet crab (Telmessus cheiragonus) (47 days) (Fisher, 2006).
Our study showed that the genetic distance values are close to the minimum genetic divergence threshold (D = 1) defined by Nei (1972), indicating no significant genetic differentiation within the blue swimming crab populations across these areas. Furthermore, an Nm value of more than 1 suggests significant migration (Slatkin, 1987). These results indicate a high level of gene flow among the crab populations in Bandon Bay. The absence of geographic barriers in Bandon Bay facilitates uninterrupted gene flow among blue swimming crab populations. The remarkable dispersal capacity of marine crab larvae frequently maintains genetic uniformity over large distances. For instance, Silva et al. (2010) investigated the population structure of Uca annulipes along the East African coast using mtDNA as a genetic marker, finding a high level of gene flow across 3,000 km of coastline. Bandon Bay’s 477 km² area and 120 km coastline allow plenty of room for blue swimming crab larvae to disperse, limiting the formation of unique population patterns. To further elucidate the genetic structure of blue swimming crabs in Bandon Bay, we recommend employing more sensitive nuclear DNA markers in future studies. Importantly, our genetic analysis revealed no differences between berried females and wild crabs from the same region. This finding suggests that the restocking program has been successful, with no evidence of genetic contamination in the population.
Our demographic history analysis revealed that the berried female and wild blue swimming crab populations in Bandon Bay maintained a stable size for an extended period before experiencing a recent expansion. Initial neutrality tests using Tajima’s D and Fu’s Fs yielded negative values, indicating a significant deviation from neutrality. This result suggests that the blue swimming crab population may have undergone either growth or purifying selection (Yang, 2006). Additionally, the negative Fu’s Fs values, which detect population expansion based on haplotype data, provide strong evidence of expansion (Fu, 1997). Furthermore, the multimodal mismatch distribution observed supports the notion of long-term stable population size. Finally, the higher θ1 compared to θ0 further corroborates the occurrence of demographic expansion. These findings align with a report that blue swimming crabs in the Gulf of Thailand had experienced a stable population phase followed by recent expansion (Supmee et al., 2020).
The crab bank project is designed to enhance wild crab populations as a countermeasure to the pressures of intensive crab fishing. Offspring from the project either grow to become breeding adults or are harvested in subsequent fishing activities. Fisheries statistics indicate an increase in blue swimming crab catches since 2019, coinciding with the Thai government’s efforts to promote the crab bank initiative (Fishery Statistics Analysis and Research Group, 2024). However, there has been limited published research on the effectiveness of stock enhancement efforts through Thailand’s crab bank project.
Stock enhancement evaluations of blue swimming crabs have been documented in several studies, including those in Australia and Japan. In Australia, anchor tags were attached to mature crabs using the mark-recapture method, but recapture rates were notably low (Williams, 1986). Similarly, a study in Japan clipped the dactyls of swimming legs from juvenile crabs and employed the mark-recapture method, with only about 4% of released crabs being recaptured (Obata, 2016). These studies highlight challenges in using traditional tagging methods for blue swimming crabs. The crabs’ molting process often leads to the detachment and loss of physical tags, while dactylus clipping may impact their behavior and survival in the wild. Furthermore, the low number of crabs released in these studies (around 10,000 individuals) likely contributed to the low recapture rates. In contrast, our study employs genetic markers from the mtDNA CR to track crab populations. This method identifies millions of progenies generated from a single-berried female crab, allowing for the release of many more crab offspring (> 1,000,000 individuals). As a result, genetic markers provide a higher detection success rate in wild crabs. The DNA-based tracking is effective and well-suited for monitoring stock enhancement, as it increases the likelihood of identifying marked individuals in the wild. However, future studies should incorporate additional genetic markers and expand sample collection to include crabs from regions beyond Bandon Bay or other nearby areas to enhance the accuracy and robustness of the tracking efforts.
Our analysis of nucleotide sequences in the mtDNA CR revealed high genetic diversity within each crab bank, indicating population fitness among blue swimming crabs in Bandon Bay. Inbreeding does not appear to be a concern for blue swimming crabs. Genetic structure analysis showed no significant genetic differentiation among crabs in Bandon Bay, suggesting that the population should be managed as a single fishery unit. This approach involves implementing stock assessments and harvest limits for the entire area while maintaining management strategies to prevent genetic contamination. For instance, crab offspring should be released only in Bandon Bay to preserve local genetic integrity. Improving coastal pollution control promotes larval dispersal and increases the number of breeding individuals. Effective management strategies should also include regulating fishing gear, monitoring habitats, and initiating restoration projects. Additionally, periodic assessments of genetic diversity and seascape studies are essential for understanding the temporal and spatial dynamics of the crab population, enabling informed and adaptive management practices.
Stock enhancement programs aim to achieve ecological, economic, and social benefits, highlighting the importance of balancing release strategies with their environmental impact. Therefore, the success of such initiatives depends not only on the survival and performance of the released recruits but also on effective fisheries management and improvements to the surrounding environment.
CONCLUSION
In this study, we analyzed 391–396 base pair mtDNA CR nucleotide sequences to assess the genetic diversity of berried and wild blue swimming crabs from crab banks in Bandon Bay. Our results revealed high genetic diversity among blue swimming crabs involved in the crab bank initiative, reflecting population fitness. Notably, 58.39% of wild crabs shared haplotypes with berried female crabs from the crab bank project, indicating that releasing crab offspring into their natural habitat contributes to the presence of adult crabs and may enhance the wild population to a sustainable level for fishing. Genetic structure analysis showed no significant differentiation within the crab population in Bandon Bay. Demographic history tests revealed a stable population size over an extended period, followed by a recent expansion. These findings provide valuable insights for developing management strategies to preserve the genetic diversity of the blue swimming crab population in Bandon Bay. Future research should incorporate nuclear DNA markers to expand the scope of genetic data and offer deeper insights into the population’s genetic dynamics.








