Introduction
The alarming situation of chloramphenicol (CAL) antibiotic resistant has become a global concern, especially in the aquatic environment. The situation is worsened by pollution from industry, agriculture, or domestic life, hence, making the aquatic environment an ideal setting for the transmission of antibiotic resistance genes in the ecosystem. Water sources are closely related to human life’s daily activities. Direct or indirect contact with water contaminated by such antibiotic resistant bacteria could harm and infect the human populace. Global dissemination of increasingly antibiotic resistant bacteria in developing countries is caused by the uncontrolled fecal disposal to surface water as mentioned by Manyi-Loh et al. (2018). The resistant gene carried by the bacteria can also migrate to microorganisms in humans via horizontal gene transfer (Bello-López et al., 2019).
CAL is a broad-spectrum antibiotic effective against both gram-positive and gram-negative bacteria (Lorenzo, 2019). It is commonly apply in aquaculture as a disinfectant or chemotherapeutic agent in treating fish diseases. The use of CAL in treating bacterial infection in food-producing animals has been prohibited in Malaysia since 1998 as stated in Malaysia Food Act 1983 (Act 281) & Regulations (Ministry of Health, 2014) as it can cause detrimental effects to human health. The European Union had ceased all aquaculture food imports from Asian countries in 2001 due to illegal CAL. CAL can cause thrombocytopenia, bone marrow depression, and aplastic anemia and produce positive results for genotoxicity in consumers (Keen & Montforts, 2012).
Shade et al. (2012) found that there is a fundamental relationship between the level of antibiotic characteristics and the distance from the disturbed stream. At the discharge site of the sewage treatment facility, it was found that an abundance of antibiotic resistant bacteria was present, but the concentration decreases gradually as the distance from the discharge site increased (Lupan et al., 2017). The water used in aquaculture is often discharged into the sea or river along with other pollutants from aquaculture farms. The contamination from sewage combined with the use of CAL will encourage the development of high resistance bacteria. If the food source is to be contaminated with CAL resistant bacteria, it may increase the risk of the microflora bacteria population inside the consumers developing antibiotic resistance. Bacteria typically develop resistance to CAL in the presence of plasmid-encoded or chromosomally encoded enzyme named chloramphenicol acetyltransferases or CAT. Huang et al. (2017) stated that CAT inactivates the effect of CAL by acetylation. The bacteria can develop CAL resistance by two separate events. Firstly, CAT catalyzes the transfer of an acetyl group from acetyl-coenzyme A to the 3-hydroxyl group of CAL (Biswas et al., 2012) to generate 3-O-acetyl-CAL, preventing it from binding to ribosomes (Huang et al., 2017). Secondly, a non-enzymatic event will occur, which begins with subsequent transfer of the acetyl group of acetyl-coenzyme A to the neighboring 1-hydroxyl group to generate 1-O-acetyl-CAL, resulting in a di-acetylated CAL (Kleanthous et al., 1984).
The CAT enzymes are divided into three types, namely cat I, cat II, and cat III. Genomic analysis of cat I, which is present in most pathogens, displays a modest sequence identical to cat II (46%) and cat III (32%–47%) (Biswas et al., 2012). A quadruple mutant of cat III named cat IV was introduced, cat IV has mutated catalytic pocket residues to mimic cat I reactions (Biswas et al., 2012). cat I was well documented and reported as a proteomic tool in a common CAL-resistance marker encoded in plasmids (Li et al., 2011a; Li et al., 2011b; Speck et al., 2011). The cat II enzyme is the only enzyme with extreme susceptibility to thiol-modifying agents compared with other cat enzymes (Murray et al., 1990) and can be found in many transposable elements (Roberts & Schwarz, 2017). The cat III enzyme is responsible for conferring resistance of the antibiotic CAL to the cell. Structural and biochemical studies of cat III were well studied to understand the general features of cat I proteins (Barsukov et al., 1996). The sequence differences between cat I, cat II, cat III, and cat IV include several substitutions in the binding site which may cause positional differences of CAL bound to these two proteins (Biswas et al., 2012). Studies have shown that E. coli bacteria are more prompt to develop antibiotic resistance if they are exposed to an increasingly high concentration of antibiotics in the surrounding environment (Tonoyan et al., 2019).
Sarawak, a state in Malaysia, is one of the most developed states. The Rajang River serves the cities on the northwest coast of Borneo. Unfortunately, the rivers and streams are dangerously polluted from the cities’ discharges (Leong et al., 2018). One of the many pollutants seeping into the waters is antibiotics via animal waste (Leong et al., 2020) or manure fertilization (Zhang et al., 2017). In Malaysia, manure is often recycled as cheap fertilizers for plantations. There are two antibiotics CAL and nitrofurans that are currently banned in Malaysia for use in agriculture since 1998 (Ministry of Health Malaysia, 2014). Farmers still illegally purchase and use CAL in most meat production (Berendsen et al., 2010). As a result, the leaching of CAL antibiotics from the fertilizer may find its way into the stream, thereby spreading into environmental bacteria (Manyi-Loh et al., 2018). Contaminants in the Food Chain (CONTAM) of European Food Security Authority (EFSA, 2014) also stated that the CAL will travel through the land system and end up contaminating the water source. In 2001, the detection of CAL in prawns happened in European countries, Australia and Asian countries such as Cambodia, Indonesia has sparked controversy because some argued that it was a huge economical waste to dispose of all the shrimp when there were only low levels of CAL found in the prawns (FAO, 2008). The use of CAL has been banned in Malaysia because it poses a serious risk to human health (Ng et al., 2014). However, the United States Food and Drug Administration (USFDA) had stopped 18 shipments from 11 Malaysian shrimp exporters due to the CAL detection in 2019 and 56 shipment of shrimp samples contained nitrofurans between 2009 and 2018 (The Straits Times, 2020). The aquaculture farms selected in the study are shrimp, prawn breeding, freshwater fish fry hatchery farms for export and the rivers are the mainstream connecting and covering the region which may be polluted by the drainage from the farms. They are the only commercial farms accessible in the area. E. coli is an opportunistic pathogen that can endure well in an aquatic system and is exceptionally proficient at horizontal gene transfer, which is considered as the vector for dissemination of antibiotic resistance (Javadi et al., 2017; Lihan et al., 2021). Odonkor & Addo (2018) have found that most antibiotic resistant E. coli can transmit their antibiotic resistance to other waterborne pathogens in drinking water. In this study, we collected water samples either from the aquaculture farms or rivers flowing from the Rajang River to 1) detect the CAL resistant gene (cat) in E. coli isolated and 2) investigate the correlation between cat gene with five common antibiotics used. We hypothesized that (1) E. coli isolates from different water sources may have different antibiotic resistance levels may detect at least one kind of cat gene and (2) there is a correlation between the CAL resistances with other antibiotics tested.
Materials and Methods
The study was conducted in three selected aquaculture farms and eight rivers in the Kuching Division, namely FW: PM Aquaculture Farms (1°28’12.9N, 110°19’51.5E), FAS: Asia Aquaculture Farms (1°36’01.2N, 110°24’29.2E), BK: FBako Aquaculture Farms (1°36’01.0N, 110°24’29.5E); RB: Bako River (1°40’48.6N, 110°22’30.8E); RBT: Bintawa River (1°35’49.5N, 110°22’30.8E), RDM: Buntal River (1°40’48.6N, 110°22’34.9E), RMT: Muara Tuang River (1°29’42.7N, 110°23’41.3E), RST: Santubong River (1°39’32.0N, 110°21’00.9E), RSN: Siniawan River (1°26’48.3N, 110°13’10.9E), RWF: Sarawak River (1°33’41.1N, 110°23’58.9E), RWC: Windcave River (1°24’52.8N, 110°08’15.2E). Sarawak is a state in Malaysia that stretches along the Northwest coastal area of Borneo. It is one of the major oils and gas and agriculture producers in Malaysia. The climate is tropical with alternate rain (October to March 2019) and dry seasons (April to September 2019). The total monthly rainfall ranged between 374–458 mm during the rainy seasons (Malaysia Meteorological Department, 2019). The water bodies receive residential waste and drainage of nearby farms along with the year-round runoff from the plantations. PM Aquaculture Farms are well-established hatchery supplying the majority of the freshwater fish fry in Sarawak; Asia Aquaculture Farms are well-established exporters for shrimp farming in Sarawak; Bako Aquaculture Farms are established exporters for freshwater fishes and prawn in Sarawak.
A total of 80 individual water samples not including triplicate were collected from each farm and river from April to September 2019 using sterile 50 mL falcon tubes. The river water samples were collected upstream from the bridge with fast-moving water. The samples were collected from the surface water and bottom (3–4 feet, 121 cm depth) of each source and placed inside an icebox to avoid adverse compositional change in the total bacterial count for further analysis in the laboratory.
The water samples were serial diluted and 20 µL of the mixture was plated on Eosin-Methylene Blue (EMB) agar (Oxoid, Basingstoke, UK) and then incubated for 24–48 hours at 28°C. Only bacterial colonies with green metallic colours were sub-cultured on new EMB agar to obtain a pure colony. The presumptive E. coli was further confirmed with indole-methyl red-Voges Proskauer-citrate (IMViC) tests which involved Sulfide-Indole-Motility (SIM), Methyl Red and Voges-Proskauer (MR-VP) and citrate tests following the standard method (Al-Baer & Hussein, 2017). The control was set using E. coli strain ATCC 25922 (Thermo Fisher Scientific, Waltham, MA, USA) and Enterobacter aerogenes (ATCC13048).
The presumptive E. coli DNAs were extracted using the boiling method as described by Sien et al. (2013) with minor modification. Total of 2 µL of DNA (10 pg) was extracted and placed directly on the spectrophotometer (NanoDrop, Thermo Fisher Scientific) for measurement at 230, 260, and 280 nm in order to assess DNA purity. The identification of the isolates was analyzed with 16S rRNA PCR by amplifying the 16S ribosomal RNA gene (Sien et al., 2013). The primers 27F 5’-AGAGTTTGATCMTGGCTCAG-3’ and 519R 5’-GWATTACCGCGGCKGCTG-3’ (Promega, Madison, WI, USA) were used in the PCR amplification. The PCR reactions (Promega) were performed in reaction mixtures containing 20 μL DNA (10 pg), 1.0 μL every 20 pmol primers, 3 μL of 10 mM deoxynucleoside triphosphates mix, 6.0 μL of 25 mM MgCl2, 10.0 μL of 5X Buffer solution, 8.0 μL of distilled water, and 1 μL of Taq polymerase. PCR was performed as follows: initial denaturation at 95°C for 10 min, denaturation at 94°C for 30 sec, annealing at 55°C for 1 min, primer extension 72°C for 1.5 min with 35 cycles and the final extension at 72°C for 10 min. Finally, the PCR products were visualized with ethidium bromide staining after gel electrophoresis in 1% agarose for 30 min at 90 V.
The 16S ribosomal DNA was purified using the Qiagen sequencing purification kit (QIAquick Gel Extraction Kit, QIAGEN, Hilden, Germany). The purified DNA product was sent to Apical Scientific Sdn. Bhd., Malaysia, for the DNA sequencing process. All 16S rRNA sequences were compared using the BLAST program to determine the closest identity matches.
The E. coli isolates were analyzed using the disk diffusion method stated in the standard procedure of the Clinical and Laboratory Standards Institute (CLSI, 2020) to evaluate its antibiotic resistance. The E. coli isolates were tested against amoxicillin (3 μg/ mL) (Oxoid), tetracycline (30 μg/ mL) (Oxoid), nitrofurantoin disk (300 μg/ mL) (Oxoid), nalidixic acid (30 μg/ mL) (Oxoid), and CAL (30 μg/ mL) (Oxoid). Reference culture (E. coli ATCC 25922) was used as a control. The test was performed in duplicate. The formation of clear zones was recorded and the susceptibility results were compared to the standard set by the CLSI (2020).
Multiplex PCR assays were used to determine the presence of CAL resistance genes in CAL-resistant E. coli. There were four sense primers c to detect the cat I, cat II, cat III, and cat IV genes. The antisense primer C-R was also added for PCR amplification. The sequence of each primer was listed in Table 1. The multiplex PCR was performed in a total volume of 25 μL using 5X Green GoTaq kit (Promega): 5 μL PCR buffer solution (5 μ/μlL), 3 μL 50 mM MgCl2, 0.8 μL of 2 mM dNTP mix, 1 μL of each primer, 0.3 μL of Taq DNA polymerase, 6.9 μL of sterile distilled water, 4 μL of crude DNA. The thermal cycling condition of multiplex PCR in cat gene amplification was started with an initial denaturation at 95°C (3 min), next by denaturation, annealing, and extension (34 cycles) at 95°C (1 min), 54°C (1 min), and 72°C (1 min), respectively. Final extension was carried out at 72°C for 5 mins. Amplification was performed in a PCR machine: Bioer Little Genius thermal-cycler (Bioer, China). A positive control (DUS-WF-6: E. coli containing cat genes obtained from Universiti Malaysia Sarawak [Ng et al., 2014]), a negative control (Pseudomonas aeruginosa ATCC 27853) and a blank control were employed in this assay. The temperature program was set according to Ng et al. (2014). The amplification products were analyzed by electrophoresis in a 1.0% (w/v) agarose at 80 V, 200 mA for 60 min. A 1kb DNA ladder (Promega) was used and the amplified fragments were visualized with a UV transilluminator.
Results and Discussion
Out of 42 bacterial colonies, 34 isolates were positive in the IMViC tests and were confirmed as E. coli. The result showed the presence of E. coli in Asia Aquaculture Farm (FAS-NS(S), FAS-SD1(S), FAS-RV1(S), FAS-R10(B), FAS-8(S), FAS-16(S), FAS-16(B), FAS-42(S), FAS-62(S), FAS-62(B)), FAS-SD2(B)), Bako Aquaculture Farm (FBK2-K4(S), FBK2-OLT(S), FBK2-OLT(B), FBK2-K2(B), FBK-RV2(S)), Bako River (RBK-S2-A), Bintawa River (RBT-S1-A, RBT-S3-A), Buntal River (RDM-S1-A, RDM-S2-A, RDM-S3-A), Muara Tuang River (RMT-S1-A, RMT-S1-B), Siniawan River (RSN-S1-A, RSN-S2-A), Windcave River (RWC-S1-A, RWC-S3-A, RWC-S3-B), Sarawak River (RWF-S1-A, RWF-S2-A, RWF-S3-A, RWF-S3-B). E. coli isolates were distributed almost equally between the aquaculture farms (n = 15) and rivers (n = 16).
The antibiotic susceptibility test of the 34 E. coli isolates against five different aqua-farming antibiotics is illustrated in Figs 1–2. The result showed that 85.3% of E. coli isolates were resistant against amoxicillin, followed by 35.3% were resistant against tetracycline, 29.4% were resistant against CAL, 17.6% were resistant against nitrofurantoin (17.6%), and 8.8% were resistant against nalidixic acid (8.8%) (Fig. 2). Most of the E. coli isolated either from farms or rivers were resistant against amoxicillin and tetracycline. Both antibiotics are broad-spectrum antibiotics commonly used to treat bacterial infections in fish. E. coli isolates that were resistant towards CAL were also resistant towards tetracycline except for FAS-16(B) and RBK-RV2(S). Among the 10 CAL resistant isolates, seven isolates were from environmental samples and another three isolates originated from aquaculture farms.
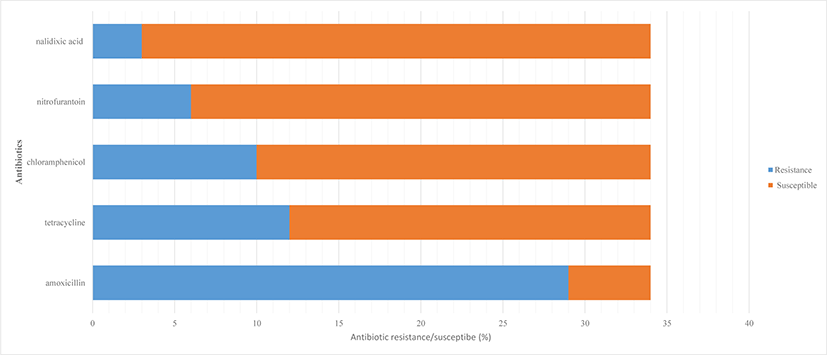
Amoxicillin was the most prevalent antibiotic detected in this study. The result was in agreement with Turnipseed et al. (2018), who detected a high level of amoxicillin residue in catfish, trout, and even in salmon farms by liquid chromatography high-resolution mass spectrometry (LC-HRMS) screening in Europe. 85.3% of amoxicillin resistant E. coli detected in the study was lower as compared to Lihan et al. (2021), who stated 100% amoxicillin resistance detected in E. coli isolated from aquaculture farms in Sarawak, Borneo. Zakaria (2017) reported that tetracycline is the second-highest antibiotic used in Malaysia up to 73,910 kg per year. This is because tetracycline antibiotics are cheap and have high efficacy against bacteria, parasites, and fungi. Tetracycline is commonly administered in water to be absorbed by the fish gills, thus the antibiotic residue is easily drained into the environment. Our result showed CAL as top three most prevalent antibiotics detected. CAL exhibits antibacterial properties by stopping the protein production in bacterial growth, thereby slowing down their infestation and multiplication (Dinos et al., 2016) whereas tetracycline is also known to inhibit bacterial protein synthesis (Chukwudi, 2016). Fig. 2 showed 6 E. coli isolated from Asia aquaculture farm and Bako Aquaculture farm was resistant to nitrofurantoin antibiotic. CAL and nitrofurantoin were banned in farming since 1998, but obviously, these two prawn and shrimp breeding farms are still applying these antibiotics in treating diseases. Most Southeast Asian countries are lacking regulatory surveillance and monitoring systems on the use of banned antibiotics (FAO & WHO, 2020). Thus, illegal usage of antibiotic cases especially in the environment, and aquaculture products are still available globally (Lai et al., 2018). Banned antibiotics such as CAL and nitrofurantoin were still detectable in our study was supported by Thiang et al. (2021), who detected tetracyclines, sulfonamides and quinolones in wastewater draining from Malaysian aquaculture farms in most Peninsular Malaysia. Thiang et al. (2021) indicated a wide distribution of antibiotics used in most aquaculture farms in Malaysia and become a resistance hotspot in promoting the growth of antibiotic resistant bacteria. Antibiotics residue in manure and other waste-based fertilizers may run off into waterways by exchanging resistance genes and altering the microbial community in water and sediment (Lihan et al., 2021). Thus, it may contribute to antibiotic resistance bacteria cases in our study. Only three E. coli were resistant to nalidixic acid (Fig. 2). Our antibiotic resistance result was higher compared to Ramírez Castillo et al. (2013), who has detected CAL (22%) and nitrofurantoin (7.7%) resistant E. coli found in rivers water in Aguascalientes, Mexico. Quinolone resistance such as nalidixic acid resistance is quite rare and resides commonly in fecal flora. Our result detected 8.8% nalidixic acid resistance E. coli, which is slightly higher compared to 13.7% resistance cases being reported in Ghana, Africa (Namboodiri et al., 2011).
In this study, we proceed to investigate the cat genes carried by the ten CAL resistant E. coli isolates. E. coli is an indicator organism that is suitable for determining CAL resistance of bacteria in Southeast Asian aquaculture environments as they are the only widespread group of CAL resistance heterotrophs that are not restricted by geographical region (Mohamed et al., 2015). E. coli is the host-specific which indicates fecal contamination in the water (Zhi et al., 2015).
The outcomes of the partial 16S rRNA sequencing homology search (BLAST) of ten E. coli isolates are shown in Table 2. The 482 genome sequences with the primer pair 27F-519R covering V1 to V3 region in 16s rRNA gene were sequenced. All the 10 isolates were identified as E. coli with 99% similarity compared to the reference genome in BLAST NCBI.
The electrophoresis result of multiplex PCR products amplified with primers C-1, C-2, C-3, C-4, and C-R are shown in Figs 3A and 3B. Fig. 3A depicts the isolates of RDM-S2-A, and RDM-S3-A detected with cat I genes compared with positive control. Fig. 3B shows all of the PCR products of E. coli that were detected with cat II genes. RSN-S1-A, RSN-S2-A and FBK2-K2 (Fig. 3A) were detected with cat III genes whereas RBK-S2-A, RDM-S1-A, RWF-S1-A, FAS-RV1(S), and FBK2-OLT(B) were detected with cat IV genes (Fig. 3B). Table 3 describes the overall CAL resistance gene detection in E. coli isolates. The results revealed that ten E. coli isolates were presented with cat II genes, two isolates were detected with cat I genes, three isolates were detected with cat III genes and five isolates were detected with cat IV genes.

Antimicrobial resistance indicators are theoretically omnipresent in the environment where the presence of such indicators is readily observed and possess genetic flexibility that allows them to obtain mobile resistance elements (Singer et al., 2016). The transferable genes encoding CAT are responsible for the antibiotic resistant characteristic of E. coli towards CAL (Munita & Arias, 2016). Furthermore, E. coli is believed to be the reservoir of other resistant genes. The resistance gene in E. coli can be transferred to other zoonotic or commensal organisms causing drug resistance in humans or cattle which can hinder standard treatment protocol (Djordjevic et al., 2013; Salinas et al., 2019).
Research has found that CAL resistance is genetically related to two different mechanisms. The first mechanism is the CAT enzymes encoded in cat genes that convert CAL into harmless molecules as discussed in this study (cat I, cat II, cat III, and cat IV genes). The second mechanism is the change in permeability to antibiotics through the efflux mechanism controlled by cmIA genes (Munita & Arias, 2016). Gene cmlA confers as non-enzymatic CAL resistance acquired via an efflux mechanism and resembles cmIA protein identified in most Pseudomonas spp. Thus, in this study cat gene is widely applied especially in CAL resistance in Gram negative bacteria (Miranda et al., 2015). For this study, all the CAL resistant E. coli isolates demonstrated two types of cat genes. The CAL resistant E. coli must exhibit at least two cat gene which encodes CAT in order to inactivate the drugs CAL by acetylation. This was proven by Martinez & Baquero (2000) that high-level resistance frequently requires mutations in more than one gene. The result was supported by Ng et al. (2014), who detected 19 E. coli isolates with cat I, 13 E. coli isolates with cat II, 10 E. coli isolates with cat III and 6 E. coli isolates with cat IV genes found in water samples of aquaculture farms in Kuching, Malaysia. Pearson correlation coefficient shows a highly significant relationship between resistance pattern of CAL with amoxicillin; and CAL with tetracycline. CAL and amoxicillin resistance occurred in E. coli isolated are significantly correlated (p = 0.026, r = 0.382, n = 34). CAL and tetracycline resistance occurred in E. coli isolated are highly significantly correlated (p = 0.001, r = 0.824, n = 34). The result was different compared to Zhang et al. (2014) who revealed significant correlations were found frequently among tetracycline, chlortetracycline, and quinolone resistance. Significant correlations between resistance to tetracycline and CAL were shown by Chen et al. (2011). These strong correlations among the top three most prevalent antibiotics may cause by co-selection (Dang et al., 2006). Studies showed that many antibiotic resistance genes are located on a similar plasmid (Carattoli, 2013), thus, these may result in the correlations found among resistance to different antibiotics.
Although low number of CAL resistant bacteria was detected among E. coli isolates found in aquaculture farms, this study further substantiates the presence of CAL in the environment. The correlation relationship of amoxicillin, tetracycline, and CAL are interrelated. The misuse of CAL as an antibiotic by local aqua-culturists despite being banned by the European Union (Tsai et al., 2019) may also trigger amoxicillin and tetracycline resistant bacteria. According to Munita & Arias (2016), the manifestation of the antibacterial resistance among bacteria that share similar properties may vary with time or period due to certain environmental and biological factors. Based on the current findings, E. coli isolated from the river samples contained a higher percentage occurrence of antibiotic resistance if compared to isolates found in aquaculture farms. Li et al. (2013) stated that a dramatic decline in temperature, pH, and dissolved oxygen in recent years, along with river runoff and sewage discharge may have caused temporal and spatial shifts in the population of antibiotic-resistant bacteria. Nonetheless, factors that determine CAL resistances in bacteria vary with environments and geological locations. Thus, monitoring the antibiotic resistance bacteria especially CAL resistance in both farms and the environment is vital before causing devastating effects to the ecosystem.
Conclusion
Total 34 of E. coli isolated from aquaculture farm and environmental water. The E. coli was resistant to amoxicillin (85.3%), tetracycline (35.3%), CAL (29.4%), nitrofurantoin (17.6%), and nalidixic acid (8.8%) antibiotics. Ten CAL resistant E. coli isolates were detected with cat II gene (100%), followed by cat IV gene (50%) and cat III gene (30%) and cat I genes (20%). All isolates contain at least two different cat genes in order to show resistance trait. Pearson correlation coefficient is showing a strong relationship between CAL with amoxicillin; and CAL with tetracycline.








